Abstract
Missing data are a recurring problem that can cause bias or lead to inefficient analyses. Development of statistical methods to address missingness have been actively pursued in recent years, including imputation, likelihood and weighting approaches. Each approach is more complicated when there are many patterns of missing values, or when both categorical and continuous random variables are involved. Implementations of routines to incorporate observations with incomplete variables in regression models are now widely available. We review these routines in the context of a motivating example from a large health services research dataset. While there are still limitations to the current implementations, and additional efforts are required of the analyst, it is feasible to incorporate partially observed values, and these methods should be utilized in practice.
Keywords: incomplete data, maximum likelihood, multiple imputation, conditional Gaussian, psychiatric epidemiology, health services research
1 Introduction
Missing data are a frequent complication of any real-world study. The causes of miss-ingness are often numerous, some due to design, and some to chance. Some variables may not be collected from all subjects, some subjects may decline to provide values, and some information may be purposely excised, for example to protect confidentiality. While the use of complete case methods that drop subjects missing any observations are commonly seen in practice, this approach has the disadvantage of being inefficient as well as potentially biased.
The development of methods for analysis of data with incomplete values has been an active area of research. Models that incorporate partially observed predictors are of particular interest in many real world settings, since missingness of just a few percent on each of a number of covariates may lead to a large number of observations with some missing information. The excellent textbooks by Little & Rubin (2002), Schafer (1997) and Allison (2002) provide a comprehensive overview of methods in this setting, focused primarily on multiple imputation. While somewhat dated, Little (1992) describes a hierarchy of approaches to account for missing predictors, including the maximum likelihood approach of Ibrahim (1990). Publications by Meng (2000) and Raghunathan (2004) provide a general introduction, while the paper by Ibrahim, Chen, Lipsitz & Herring (2005) reviews recent developments in a comprehensive fashion, though their application (cancer dataset) features incompleteness on only one variable. A useful online annotated bibliography provides a comprehensive reading list (Carpenter 2006a).
In this article, we update the prior review of Horton & Lipsitz (2001) and apply methods described by Ibrahim et al. (2005) to the logistic regression analysis of a dataset with incompleteness on four variables (both categorical and continuous) using a variety of software packages. We discuss modeling assumptions, approaches and compromises required for estimation within current implementations. In section 2, we briefly review methods for incorporating incomplete observations in regression models then summarize findings of two surveys of how missing data methods are used in practice in section 3. We describe our motivating example (which features a large dataset with high proportion of missing values with a non-monotone pattern) in section 4, detail support for missing data software in section 5, then apply these methods to the motivating dataset in section 6. We contrast the strengths and limitations of these packages in practice, and suggest improvements for the future.
We focus on methods to incorporate partially observed predictors; different issues arise when some outcomes are not fully observed. We also do not consider longitudinal or clustered outcomes, for which other complications arise (e.g. Laird (1988), Robins, Rotnitzky & Zhao (1995) and Jansen, Beunckens, Molenberghs, Verbeke & Mallinckrodt (2006)).
2 Incomplete data regression methods
2.1 Notation and nomenclature
We begin by introducing notation that will be used throughout, assuming that data are collected on a sample of n subjects and that primary interest relates to the parameters governing the conditional distribution f(Yi|Xi,β). To simplify exposition, we suppress the subject indicator. For a given subject we can partition X into components denoting observed variables (Xobs) and those that are missing for that subject (Xmis). We denote by R a set of response indicators (i.e. Rj = 1 if the jth element of X is observed, and equals 0 otherwise), governed by parameters φ. Little & Rubin (2002) introduced a nomenclature for missingness in terms of probability models for R. The missing completely at random (MCAR) assumption is defined as
where in addition φ and β are presumed distinct. Note that this depends on an assumed relationship between R and the unobserved and unknowable Xmis. Heuristically, this assumption states that missingness is not related to any factor, known or unknown, in the study.
It may be more plausible to posit that missingness is missing at random (MAR), which assumes that
This assumption states that the missingness depends only on observed quantities, which may include outcomes and predictors (in which case the missingness is sometimes labeled covariate dependent missingness (CDM)). The assumption regarding the lack of association with unobserved quantities (Xmis) remains. We note that at first glance, the meaning of the term “missing at random” can be confusing, since missingness can actually be predicted (but is random after controlling for missingness due to observed quantities).
Researchers have noted that by including a relatively rich set of predictors in the model, the MAR assumption may be made more plausible (Collins, Schafer & Kam 2001). Others have noted that incorporation of information regarding the outcome improves estimation of missing predictors (Moons, Donders, Stijnen & Harrell 2006).
It is possible to formally test the MCAR assumption against the alternate hypothesis of MAR (Little 1988, Diggle, Heagerty, Liang & Zeger 2002).
Finally, if the missingness law P(R|Y, X) cannot be simplified (i.e. it depends on unobserved quantities), the process is termed non-ignorable (abbreviated NINR [non-ignorable nonresponse or MNAR [missing not at random]). In a NINR setting, the correct specification of the missingness law must be given to yield consistent estimates of the regression parameters. Researchers have addressed issues in the NINR setting (Diggle & Kenward 1994, Little 1994). NINR models are particularly useful in assessing the sensitivity of results to deviations from MAR missingness (Carpenter, Kenward & Vansteelandt 2006). Without additional information, it is inherently impossible to test whether MAR holds (Little & Rubin 1987).
Another important concept regarding missing data, particularly where there are multiple variables with missing values, relates to the pattern of missing data. If the data matrix can be rearranged in such a way that there is a hierarchy of missingness, so that observing a particular variable Xb for a subject implies that Xa is observed, for a < b, then the missingness is said to be monotone. Figure 1 displays two hypothetical datasets. The left hand dataset has been rearranged to create a monotone ('stair-step') pattern, though this is not possible in the right hand side of the figure. When missingness is non-monotone, models for the missingness of one variable may include covariates which are also missing values. Simpler methods can be utilized if the pattern is monotone, though a monotone pattern is uncommon in most realistic settings (including our motivating example).
Figure 1.
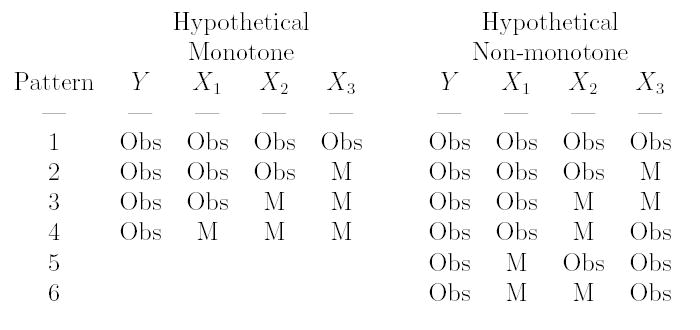
Monotone and non-monotone patterns of missingness (Obs=observed, M=missing)
2.2 Complete case method
The simplest method for the analysis of incomplete data regression models involves the analysis of the set of observations with no missing values. When missingness is MCAR, then the complete case (CC) estimator is unbiased. The main drawback of the CC estimator is that if there are many different variables with missing values, then a large fraction of the observations may be dropped. For the example datasets in Figure 1, only data from pattern 1 is included, though partial information is available from the other patterns (e.g. the joint distribution of f(Y, X1, X2) can be estimated from pattern 2). The efficiency losses of the complete case estimator can be substantial (Little & Rubin 2002) (p. 42).
2.3 “Ad-hoc” methods
A series of “ad-hoc” methods have been suggested to address missing data. One approach for continuous variables involves recoding missing values to some common value, creation of an indicator of missingness as a new variable, and including both these variables along with their interaction in the regression model. A similar approach for categorical variables involves the creation of an additional category for missing values. These ad-hoc approaches have the potential to induce bias and are not recommended (Jones 1996, Greenland & Finkle 1995).
A second approach involves dropping variables from the analysis that have a large proportion of missing values. This is not attractive, since it may lead to the exclusion of important factors in the regression model, with consequent bias or unnecessarily large standard errors.
Two other approaches involve imputation of missing values using mean imputation (average of the observed values) or for longitudinal studies, the last observed value (also known as last value carried forward, LVCF or last observation carried forward, LOCF). Both methods have the potential of inducing bias as well as understating variability and are not recommended (Carpenter, Kenward, Evans & White 2004, Cook, Zeng & Yi 2004, Jansen et al. 2006).
2.4 Multiple imputation
Multiple imputation is a three-step approach to estimation of incomplete data regression models due to Rubin (1976). First, plausible values for missing observations are created that reflect uncertainty about the non-response model. These values are used to “fill-in” or impute the missing values (using an assumption of MAR). This process is repeated, resulting in the creation of a number of “completed” datasets. Second, each of these datasets is analyzed using complete-data methods. Finally, the results are combined, which allows the uncertainty regarding the imputation to be taken into account. Typically 5–10 imputations are created, though more are computationally feasible and better characterize the variability introduced into the results due to the imputation process.
The method of multiple imputation was first proposed in a public use survey data setting. Multiple imputation remains ideally suited to this setting, since the creators of the data set can utilize auxiliary confidential and detailed information that would be inappropriate to include in the public dataset (Rubin 1996). Given the completed datasets, users may utilize existing software to analyse each of the datasets. Given the results for each analysis, an overall summary is straightforward to calculate.
Multiple imputation is however more commonly used in a setting where the imputer and the analyst are the same person. An extensive literature on the topic exists (Rubin 1987, Glynn, Laird & Rubin 1993, Rubin 1996, Schafer 1999, Barnard & Meng 1999, van Buuren, Boshuizen & Knook 1999, Allison 2000, Kenward & Carpenter In press) as do implementations in general purpose statistical software. A useful guide to software implementations can be found at the multiple-imputation.com website (van Buuren 2006).
The key issue for the analyst is the appropriate specification of the imputation model, since if this is misspecified, there is the potential for bias. Often a multivariate normal model has been used, as it is computationally tractable (since only the mean vector and variance-covariance matrix needs to be estimated). This model has been used even when some of the variables are not Gaussian, though this complicates analyses and if imputed values are rounded, can lead to bias (Horton, Lipsitz & Parzen 2003, Allison 2005, Bernaards, Belin & Schafer In press). These issues are particularly salient when multiple categorical and continuous variables have missing values, as the joint distribution may be complicated. Finally, note that the analysis should not use a richer model than that used for imputation (Little & Rubin 2002).
We will now review a number of methods that have been proposed for imputation models for categorical and continuous variables.
Conditional Gaussian
One approach to imputation when there are both continuous and discrete missing values is the Conditional Gaussian approach, popularized by Schafer (1997). A log-linear model (Bishop, Fienberg & Holland 1975) is specified for the discrete random variables, and conditional on this distribution, a multivariate normal distribution is assumed for the continuous variables. This general location model (Olkin & Tate 1961) can be fit as a saturated multinomial with separate means and shared covariance, but this may lead to a proliferation of parameters in real-world applications with multiple categorical variables. As a result, simplification of the log-linear model is required in practice. This approach has been implemented in the MIX programs of Schafer (assuming a form of monotonicity) as well as S-Plus missing data library.
Chained equations
An alternative approach involves a variable by variable approach using chained equations (van Buuren et al. 1999, Raghunathan, Lepkowski, van Hoewyk & Solenberger 2001, van Buuren, Brand, Groothius-Oudshoorn & Rubin 2006, van Buuren In press). The imputation model is specified separately for each variable, involving the other variables as predictors. At each stage of the algorithm, an imputation is generated for the missing variable, then this imputed value is used in the imputation of the next variable. This process repeats, imputing missing values using a Gibbs sampling procedure until the process reaches convergence. Separate chains are used to generate the multiple imputations.
For continuous variables, the model may involve a linear regression model or use predictive mean matching (where the imputed variables takes on the value of one of a set of nearest observed value in the dataset). For dichotomous variables, logistic regression can be fit, while polytomous models are needed for categorical variables. Implementations of the chained equation approach are available in the MICE library (for R and S-Plus), ICE (for Stata), IVEware (for SAS or standalone) or aregImpute (for R and S-Plus).
One problem with the chained equation approach is that it may not converge to a sensible stationary distribution if the separate models are not compatible with a multivariate distribution (Raghunathan et al. 2001), though van Buuren et al. (2006) show in a series of simulation studies that reasonable imputations were obtained even when the separate models were incompatible. Additional work is needed to further establish the validity of the approach.
Methods for monotone datasets
A number of approaches are implemented in SAS PROC MI for datasets with monotone missing structure. The predictive mean matching method can be used to impute a value randomly from a set of observed values whose predicted values are closest to the predicted value from a specified regression model. This process is straightforward when imputing a continuous random variable, but more complicated when imputing a categorical variable with more than 2 levels. The process begins with the observations with only one missing value, and then uses those values in the imputation of the observations with two missing values, etc. Analysts are warned that Allison (2000) found that predictive mean matching approaches led to biased results when applied to missing predictor models.
Similarly, regression or propensity score models can be used to impute missing values.
One disadvantage of these approaches is that for datasets with non-monotone missingness, some observations need to be dropped from the analysis, or some “ad-hoc” procedure used. As an example, consider the hypothetical non-monotone patterns in Figure 1. It is possible to create a monotone dataset by only including patterns (1,2,3), (1,3,6), (1,3,4) or (1,5,6). In practice, however, in addition to being highly arbitrary, creation of a dataset with monotone missingness may exclude a large number of observations and be inefficient as well as potentially biased.
The analyst may be able to creatively exploit patterns of missing values in a particular dataset. Consider, for example, using patterns (1,2,3) to estimate f(X2, X3|Y, X1), and using patterns (1,4) to estimate f(X2|Y, X1, X3). We pursue this type of strategy to avoid dropping observations in our motivating example.
Other issues with imputation
There are many additional imputation issues that are beyond the scope of this manuscript. For example, there may be bounds on imputed values which need to accounted for (e.g. as only zero values for years smoking are plausible for a non-smoker) (Raghunathan et al. 2001). Similar issues arise when missing values are known to be within a certain range (e.g. between 3 and 4 on a 5 point Likert scale), or when variables require transformations. While important issues, we do not further consider them.
2.5 Likelihood based approaches
Maximum likelihood is an alternative approach which also assumes that missingness is MAR. Typically, primary interest relates to the regression parameters governing the conditional distribution: f(Y |X, β). When some of the predictors are missing, however, Ibrahim (1990) suggested that information can be reclaimed by estimating the distribution of the covariates: f(X|γ). The joint distribution f(X, Y |β, γ) is maximized, typically through use of the EM (Expectation-Maximization) algorithm (Dempster, Laird & Rubin 1977). For each observation with missing data, multiple entries are created in an augmented dataset for each possible value of the missing covariates, and a probability of observing that value is estimated given the observed data and current parameter estimates (E-step). The augmented complete-data dataset can then be used to fit the regression model, accounting for these weights. Figure 2 displays a hypothetical dataset with a completely observed outcome (Y ) and three dichotomous covariates that are sometimes missing (X1, X2 and X3). Observation 3 is missing X3, so two entries are created in the augmented dataset, with w31 + w32 = 1, w31 = P (X3 = 0|Y = 1, X1 = 1, X2 = 0, β, γ). For observation 7, there are 8 entries in the augmented dataset. Horton & Laird (1999) review this methodology in detail, while Ibrahim et al. (2005) compare and contrast it with other approaches.
Figure 2.
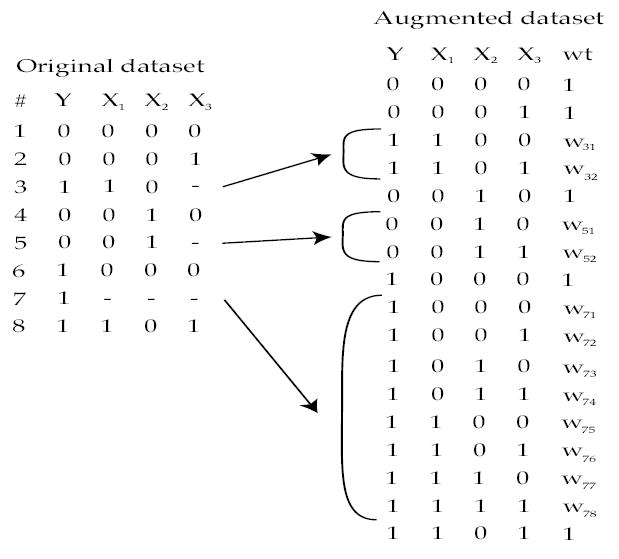
Use of Likelihood based approach with EM algorithm to incorporate partially
One of the complications of this method is the need to model the nuisance distribution of the covariates. In some settings with only a few categorical variables a saturated multinomial distribution can be fit. When there are more variables, some simplification of the joint distribution is often necessary. Lipsitz & Ibrahim (1996) suggested a conditional approach where:
where each of the marginal models typically involve only main effects. Further complications arise with continuous covariates, since some form of MCEM (Monte Carlo EM) is required (Ibrahim, Chen & Lipsitz 1999).
Another complication for maximum likelihood relates to the calculation of the standard errors of estimates. Implementations of maximum likelihood that address these complications are available in LogXact version 7, the S-Plus missing data library and in SPSS (von Hippel 2004).
2.6 Weighting methods
Another approach to accounting for missing predictor data is the use of weighting methods (Robins et al. 1995, Xie & Paik 1997, Horton & Lipsitz 1999, Horton, Laird, Murphy, Monson, Sobol & Leighton 2001, Carpenter et al. 2006). In this approach, a model for the probability of missingness is fit, and the inverse of these probabilities are used as weights for the complete cases. Weighting approaches can be fit in software that allows for weights (e.g. Stata, SUDAAN or SAS). This approach is detailed for a single missing predictor in Ibrahim et al. (2005) and Carpenter et al. (2006), but becomes considerably less tractable with multiple missing variables, particularly when they are non-monotone. Due to this limitation, and the fact that our motivating example is decidedly non-monotone, we do not further pursue estimation using weighting methods.
2.7 Bayesian approaches
While multiple imputation was derived from within a Bayesian framework (sampling from the posterior distribution of interest), Bayesian approaches have been applied more generally. Ibrahim et al. (2005) describe estimation with a prior distribution on the covariates, and the close relationship between the Bayesian approach and ML and MI methods. In part because these methods are so flexible, specific coding of prior distributions and model relationships with a package such as WinBugs is required for estimation. Such coding is relatively straightforward, and examples of missing data models are available (Carpenter, Pocock & Lamm 2002, Carpenter 2006b), however we do not further discuss these approaches.
3 Surveys of missing data methods use in practice
Despite the existence of principled methods for the analysis of incomplete data regression models, there is some evidence that their use in applied settings remains limited. We base this statement on two recent studies of statistical methods used in medical research.
Burton & Altman (2004) reviewed the reporting of missing covariate data in 100 cancer prognostic studies published in 2002. Extensive detective work by the authors determined that 81% of the articles had missing data (though the status was unclear for 4 articles, and 13 had availability of data as an inclusion criteria). Of the 81 articles with missing data, 32 stated methods for the analysis of incomplete observations (several articles used more than one method). A total of 12 papers used a complete case approach, 12 available case, 6 omitted between one and four variables, 4 used a missing indicator approach, 3 used an ad-hoc single imputation procedure, and only one paper used multiple imputation. In responses to these limitations, they proposed a set of guidelines for the reporting of studies with missing covariate data (Figure 3). In closing, Burton & Altman (2004) noted that:
Figure 3.
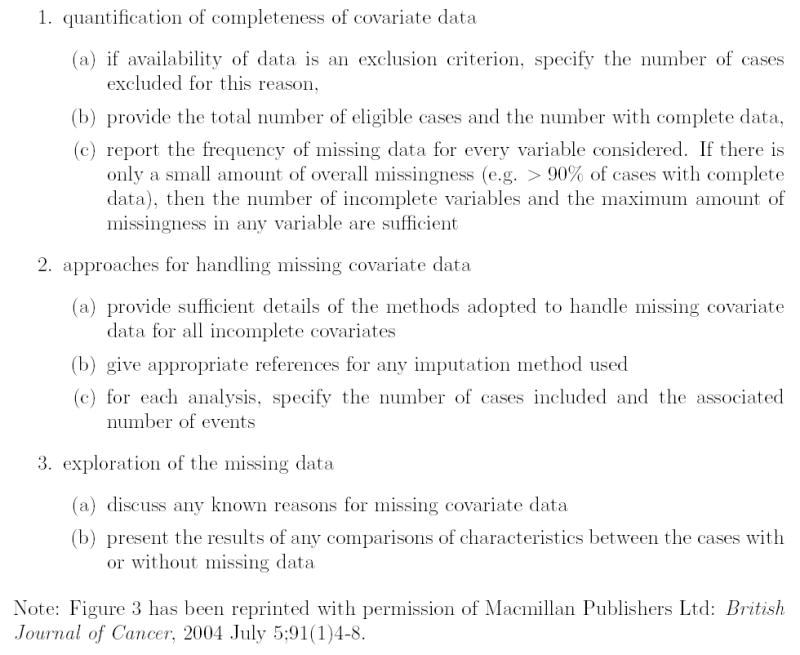
Proposed guidelines for reporting missing covariate data (Burton and Altman 2004)
We are concerned that very few authors have considered the impact of missing covariate data; it seems that missing data is generally either not recognised as an issue or considered a nuisance that is best hidden.(p.6)
Horton & Switzer (2005) reviewed the use of statistical methods in original research articles published during an 18 month period of 2004–2005 in The New England Journal of Medicine, a widely read and highly cited medical journal. Of the n=331 papers that were reviewed, 26 (8%) reported some form of missing data methods. For the purposes of this paper we further reviewed those 26 manuscripts, finding that 12 utilized a variant of last value carried forward, 13 used an ad-hoc imputation strategy (e.g. mean imputation), and 2 undertook sensitivity analyses where missing values were replaced by worst case values. Two papers used multiple imputation (Smith, Wood, Pell, White, Crossley & Dobbie 2004, van de Beek, de Gans, Spanjaard, Weisfelt, Reitsma & Vermeulen 2004). The entire description of this strategy in Smith et al. (2004) was that “missing values were estimated by multiple multivariate imputation” and a citation was given to the MICE approach of van Buuren et al. (1999). The paper by van de Beek et al. (2004) noted that only 320 of the 696 patients had complete data, and all predictors were used to impute missing values (using a multivariate normal model, though some of the variables were categorical). Neither of these two papers provided the information suggested by Burton and Altman that would be sufficient to replicate this analytic approach.
Both reviews indicate that there is a considerable gap between statistical methodologies and methods that are commonly used in practice. Flexible comprehensive implementations of these methods may spur their use.
4 Motivating example: Kids' Inpatient Database
These methods are demonstrated using the Healthcare Cost and Utilization Project (HCUP) KIDS' Inpatient Database (KID) for the year 2000 (HCUP Kids' Inpatient Database (KID) 2000). This dataset, which is publicly available for a fee from the Agency for Healthcare Quality and Research, collects data from states on child hospitalizations to improve the quality of health care. We investigated what factors predicted whether a pediatric subject with a psychiatric or substance abuse diagnosis had a routine discharge from the hospital.
More specifically, we included all 10–20 year-old subjects with a Clinical Classifications Software (CCS) category for primary, secondary or tertiary diagnosis equal to (66) alcohol-related mental disorders, (67) substance-related mental disorders, (68) senility and organic mental disorders, (69) affective disorders, (70) schizophrenia and related disorders, (71) other psychoses, (72) anxiety; somatoform; dissociative; and personality disorders; (73) pre-adult disorders, (74) other mental conditions, or (75) personal history of mental disorder; mental and behavioral problems; observation and screening for mental condition.
The outcome in our model was routine discharge vs. non-routine discharge (including transfer to a short term hospital, other facility, release to home health care, dying in hospital or leaving against medical advice).
Predictors in the logistic regression included an indicator of gender (FEMALE, 1=female, 0=male), AGE (in years), length of stay (LOS, in days), admission type (ATYPE, 1=emergency, 2=urgent, 3=elective), admission month (AMONTH, used to derive season of admission, NSEASON), admission on weekend (WEEKEND, 1=Saturday or Sunday, 0=otherwise), number of diagnoses on original record (NDX), race/ethnicity (RACE, 1=white, 2=black, 3=hispanic, 4=other), and total charges (TOTCHG, in dollars).
4.1 Descriptive statistics
Table 1 provides descriptive statistics for the observed data from the KID dataset. More than four-fifths of the sample were discharged in a routine fashion, one-fifth during the weekend, with more than half female and two-thirds white/caucasian. The average age was 16 years, and the length of stay, total charges and number of diagnoses were all skewed to the right.
Table 1.
Descriptive statistics for KID dataset
| VARIABLE | PROPORTION |
|---|---|
| routine discharge | 85.8% |
| weekend | 19.7% |
| female | 53.7% |
| RACE/ETHNICITY: white
black hispanic other |
68.4%
16.0% 10.3% 5.3% |
| SEASON: winter
spring summer fall |
23.7%
27.1% 22.9% 26.3% |
| ADMISSION TYPE: emergency
urgent elective |
50.6%
33.1% 16.3% |
| VARIABLE | MEAN (SD) |
| age (in years)
length of stay (LOS, in days) total charges (TOTCHG) number of diagnoses (NDX) |
16.3 (2.7)
6.4 (12.7) $9,230 ($17,371) 3.5 (2.0) |
4.2 Missing data
A total of 133,774 observations were recorded. Data were complete for the ROUTINE, FEMALE, AGE, LOS, WEEKEND and NDX variables.
There were missing values for TOTCHG (4% of dataset), ATYPE (11% of dataset), RACE (16% of dataset) and NSEASON (12% of dataset). AMONTH and ATYPE were missing by design since some states restrict the availability of information to minimize the possibility of inadvertent reidentification of subjects in smaller hospitals, while some states prohibited reporting data on RACE. A total of 79,574 (59%) of observations had complete data.
Because LogXact requires variables with missing values to have no more than 5 levels (coded 0, 1, . . . , 4), the variable AMONTH was recoded into a variable ASEASON where Winter was defined as months December, January or February, Spring as months March, April or May, etc.
Figure 4 displays the pattern of missing data using routines within Stata; a similar presentation can be created with SAS, R or S-Plus.
Figure 4.
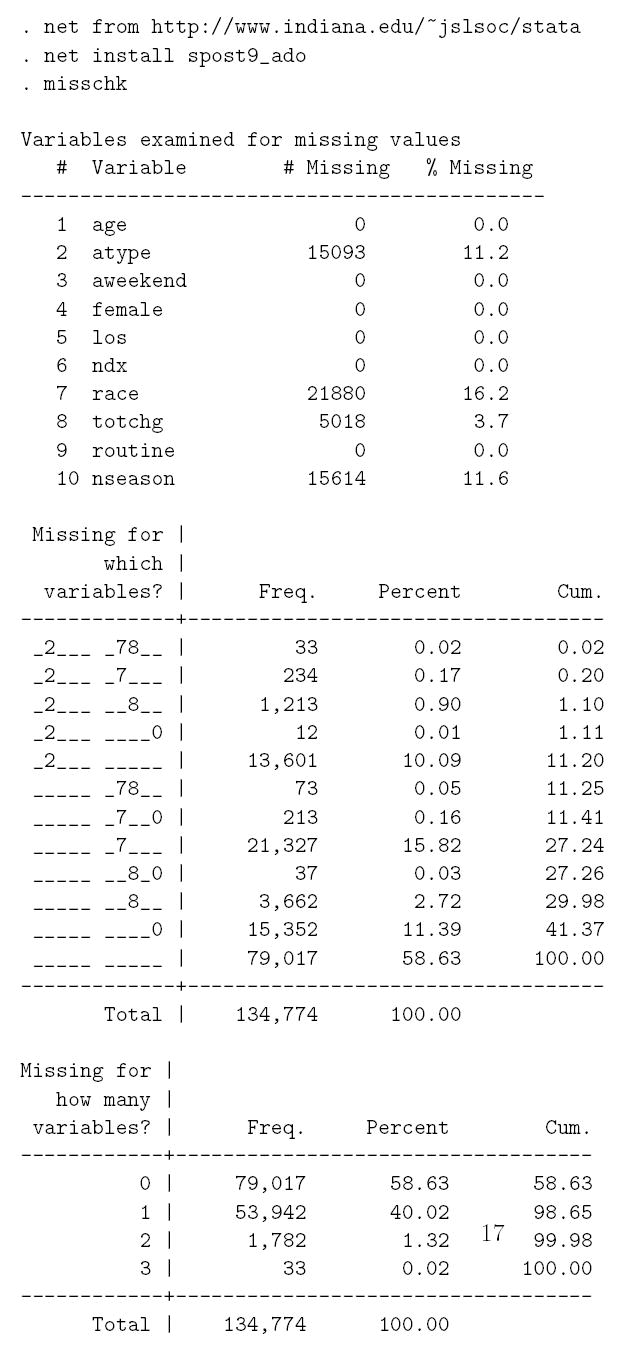
Description of missing data (using Stata misschk function)
4.3 Results
Table 2 displays the results from the complete case estimator (n=79,017). However, the use of the complete case estimator means that incomplete observations are excluded from the analysis, even though for almost all subjects, complete data on the outcome as well as all but one or two predictors are available. We now review software implementations to incorporate these incomplete observations.
Table 2.
Results from complete case estimator (Stata)
| . logistic routine age nseas1 nseas2 nseas3 a2 a3 los totchg3 ndx | aweekend r2 r3 r4 female | |||||
| Logistic regression | Number of obs = 79017 | |||||
| LR chi2(14) = 1100.68 | Prob > chi2 = 0.0000 | |||||
| Log likelihood = −30802.402 | Pseudo R2 = 0.0176 | |||||
|
| ||||||
| routine | Odds Ratio | Std. Err. | z | P>|z| | [95% Conf. Interval] | |
|
| ||||||
| age | .9603541 | .0038511 | −10.09 | 0.000 | .9528357 | .9679318 |
| nseas1 | .935468 | .0278596 | −2.24 | 0.025 | .8824273 | .9916969 |
| nseas2 | .9818723 | .0302025 | −0.59 | 0.552 | .9244255 | 1.042889 |
| nseas3 | 1.038108 | .0308933 | 1.26 | 0.209 | .9792904 | 1.100459 |
| aweekend | .9437081 | .0246635 | −2.22 | 0.027 | .8965856 | .9933072 |
| r2 | .9825766 | .0286533 | −0.60 | 0.547 | .9279919 | 1.040372 |
| r3 | .8133424 | .0320256 | −5.25 | 0.000 | .7529342 | .878597 |
| r4 | .8782054 | .039837 | −2.86 | 0.004 | .8034966 | .9598607 |
| female | 1.093442 | .0230012 | 4.25 | 0.000 | 1.049277 | 1.139465 |
| a2 | 1.401183 | .0343642 | 13.75 | 0.000 | 1.335424 | 1.470181 |
| a3 | 1.474421 | .0478478 | 11.96 | 0.000 | 1.383561 | 1.571248 |
| los | .9960628 | .0008627 | −4.55 | 0.000 | .9943734 | .9977551 |
| totchg3 | .9751805 | .0067648 | −3.62 | 0.000 | .9620116 | .9885298 |
| ndx | .8979635 | .0044983 | −21.48 | 0.000 | .8891901 | .9068234 |
|
| ||||||
| . logit | ||||||
| Logistic regression | Number of obs = 79017 | |||||
| LR chi2(14) = 1100.68 | Prob > chi2 = 0.0000 | |||||
| Log likelihood = −30802.402 | Pseudo R2 = 0.0176 | |||||
|
| ||||||
| routine | Coef. | Std. Err. | z | P>|z| | [95% Conf. Interval] | |
|
| ||||||
| age | −.0404532 | .0040101 | −10.09 | 0.000 | −.0483128 | −.0325936 |
| nseas1 | −.0667083 | .0297815 | −2.24 | 0.025 | −.1250789 | −.0083377 |
| nseas2 | −.018294 | .0307601 | −0.59 | 0.552 | −.0785828 | .0419947 |
| nseas3 | .0374 | .0297592 | 1.26 | 0.209 | −.020927 | .0957271 |
| aweekend | −.0579384 | .0261347 | −2.22 | 0.027 | −.1091615 | −.0067153 |
| r2 | −.017577 | .0291614 | −0.60 | 0.547 | −.0747323 | .0395783 |
| r3 | −.2066032 | .0393753 | −5.25 | 0.000 | −.2837774 | −.1294289 |
| r4 | −.1298747 | .0453619 | −2.86 | 0.004 | −.2187824 | −.0409671 |
| female | .0893302 | .0210356 | 4.25 | 0.000 | .0481013 | .1305592 |
| a2 | .3373171 | .0245251 | 13.75 | 0.000 | .2892488 | .3853855 |
| a3 | .3882654 | .0324519 | 11.96 | 0.000 | .3246608 | .45187 |
| los | −.003945 | .0008661 | −4.55 | 0.000 | −.0056425 | −.0022475 |
| totchg3 | −.0251326 | .0069369 | −3.62 | 0.000 | −.0387288 | −.0115365 |
| ndx | −.1076259 | .0050095 | −21.48 | 0.000 | −.1174442 | −.0978075 |
| _cons | 2.803206 | .0730586 | 38.37 | 0.000 | 2.660013 | 2.946398 |
5 Software packages
Table 3 lists the missing data implementations considered in this review. For each package, we provide a general introduction and discuss any particular issues related to the implementation, strengths or limitations. For each package, we have included the code to analyse the motivating dataset along with the relevant output (Appendix).
Table 3.
General purpose software implementations of missing data routines
| ROUTINE | SOFTWARE PACKAGE | VENDOR/AUTHOR | APPROACHES IMPLEMENTED |
|---|---|---|---|
| Amelia II | R | Honaker, King and Blackwell | hybrid EM with bootstrap |
| Hmisc | R and S-Plus | Frank Harrell | chained-equation using predicted mean matching or regression imputation |
| ICE | Stata | Patrick Royston | chained-equation |
| IVEware | SAS or standalone executable | University of Michigan | chained-equation (supports complex survey designs + constraints) |
| LogXact | LogXact 7 | Cytel | Maximum likelihood |
| MICE | R and S-Plus | van Buuren et al | chained-equation (and potential NINR models) |
| PROC MI | SAS v9.1 | SAS Institute | MCMC for Gaussian, PMM, regression, logistic, polytomous and discriminant models |
| missing data library | S-Plus 7 | Insightful | Maximum likelihood or conditional Gaussian imputation model |
5.1 Amelia II
Amelia II (Honaker, King & Blackwell 2006) utilizes a bootstrapping-based EM algorithm (e.g. EMis (King, Honaker, Joseph & Scheve 2001)) that is both fast and robust. It includes features for imputing cross-sectional surveys, time series data, and time-series/cross-sectional data. The package allows users to put priors on individual missing cell values in the data matrix, when that knowledge is available. The paper by Honaker & King (2006) provides additional description of the package.
Amelia II performs the imputation step. Separate analyses and combination of results can either be undertaken in R using the Zelig (Imai, King & Lau 2006) software, or in a separate statistics package (e.g. SAS or Stata). The code to install Amelia and Zelig within R or S-Plus can be found in Figure 5 (Appendix), along with the code to combine the multiple imputations. A screenshot of AmeliaView() can be found in Figure 7 (Appendix) while the output is displayed in Figure 8 (Appendix).
In addition, a self-install package is available which allows a user to install Amelia II without any knowledge of, or even directly running, R. If this route it preferred, Amelia can output data sets for analysis and combination in another package. Figure 6 (Appendix) displays the SAS code to read (lines 1–19), analyse (lines 21–34) and combine (lines 30–40) output datasets from Amelia; more information on SAS can be found in Section 5.7.
5.2 Hmisc
The aregImpute function with the HMisc package (Harrell 2006) for R and S-Plus supports predictive mean matching with optional weighted probability sampling from similar cases. Predictive mean matching works for binary, categorical, and continuous variables, without the need for computing residuals or constraining imputed values to be in the range of observed values. This approach takes all aspects of uncertainty into account by using the bootstrap to approximate the full Bayesian predictive distribution for imputations. There is also support for regression imputation within HMisc.
The packages is available for download from CRAN (the Comprehensive R Archive Network). Figure 9 (Appendix) displays the code to read in the dataset (lines 1–12), create graphical displays (lines 13–15), impute using aregImpute (lines 17–20) then assess convergence and combine results (lines 21–28). Figure 10 displays the output from aregImpute and Figure 11 (Appendix) a series of graphical displays of missing data patterns (from Hmisc).
5.3 ICE/Stata
Patrick Royston's ICE (imputation using chained equations), implemented within Stata, provides support for categorical missing values (Royston 2005). Binary variables are predicted from other variables using logistic regression, while categorical variables with more levels using either a multinomial or ordered logistic regression. Some housekeeping is needed when using indicator variables to represent the multiple levels of a categorical variable. Figure 12 (Appendix) displays the code to analyse the KID dataset using ICE.
The software can be installed over the internet from within Stata (lines 1–2). Much of the syntax involves the creation of indicator variables with appropriate missingness structure (lines 6–24). This must be done in advance so that ice can create them from imputed values using the passive and substitute statements (lines 32–35). To allow commands to be split onto separate lines, the delimit statement is used (lines 28 and 36). Multinomial logit models are used for the SEASON, RACE and ATYPE variables, while a linear regression model is used for total charges (line 31). The imputed datasets are saved into a dataset called imputed and this is used to fit the regression model of interest (lines 40–41). Figures 13 and 14 (Appendix) display the output.
5.4 IVEWARE
IVEware (http://www.isr.umich.edu/src/smp/ive) by Raghunathan et al. is a SAS version 9 callable routine built using the SAS macro language or a standalone executable. In addition to supporting chained equations, it extends multiple imputation to support complex survey sample designs.
Installation is straightforward, and consists of 19 SAS command files. The distribution includes an example dataset to help verify the installation. Figure 15 (Appendix) displays the code to fit the logistic regression imputation model while Figure 16 (Appendix) provides the output.
5.5 LogXact
LogXact is a stand-alone package for the analysis of generalized linear models. LogXact version 7 incorporates support for the likelihood methodology of Ibrahim (1990) for up to ten categorical covariates with missing values (each with up to five levels). There is no practical limit on the number of fully observed predictors. The software functions via a graphical interface. Since our motivating example had a predictor (TOTCHG, or total charges) that was continuous, we had to drop observations where TOTCHG was not observed (approximately 4% of the dataset). One of our missing predictors was month of admission; because only 5 levels were allowed we coded this into a SEASON variable with four levels. Figure 17 (Appendix) displays a screenshot of LogXact's results from the missing data model.
We ran into some precision or collinearity issues related to the range of TOTCHG (when this variable was divided by 1,000 the models worked, as well as when we fit the untransformed variable using only complete cases). There was also a minor bug in the display routines (Cytel reports that these bugs have been fixed and will ship in the next release). The requirement that categorical variables take on values starting from zero in sequence to a max of four required some tedious recoding.
While there was an option to display the variance-covariance matrix of the regression parameters, there were fatal memory errors on the testing machine when these were checked. Access to the variance covariance matrix is needed to calculate functions of the regression parameters, or to conduct multiple-df tests.
5.6 MICE
MICE (Multiple Imputation by Chained Equations) is a library for S-Plus and R. The package is available from the Comprehensive R Archive Network. A variety of imputation models are supported, including forms of predictive mean matching and regression methods, logistic and polytomous regression and discriminant analysis. In addition, MICE allows users to program their own imputation functions. In theory, this could facilitate sensitivity analyses of different (possibly non-ignorable) missingness models. The imputation step is carried out using the mice() function.
We note that a bug initially yielded incorrect inferences from the logistic regression model. A patch is now available for release 1.14. Figure 18 (Appendix) displays the code from mice for reading in the dataset (lines 1–9), imputation (lines 10–11), and combination of results (lines 12–14). Figure 19 (Appendix) displays the output and results.
5.7 SAS PROC MI
Analysis using multiple imputation in SAS/STAT is carried out in three steps. First, the imputation is carried out by PROC MI. Then, complete data methods are employed using any of the SAS procedures for complete data analysis (e.g. PROC GLM, GENMOD, PHREG, or LOGISTIC); the 'BY' statement repeats the analysis for each completed data set. Finally, the results are combined using PROC MIANALYZE. No additional installation was needed for PROC MI/PROC MIANALYZE, since it is part of the SAS/STAT product.
PROC MI incorporates a number of different imputation methods. For non-monotone missing data patterns, the MCMC statement can be used either for all missing data or to impute enough data so that the remaining missing data is monotone. At present only continuous variables (those not specified in the CLASS statement) can be included for MCMC imputation. Extensive control and graphical diagnostics of the MCMC methods are provided.
For monotone data, the MONOTONE statement is used to describe the imputation method. Continuous variables can be imputed using multivariate regression, regression using predictive mean matching (each of these assuming normality), or via propensity scores. Categorical variables, specified in the CLASS statement, can be imputed via logistic (or ordinal logistic) regression, discriminant analysis (only continuous predictors allowed) or propensity scores. The approaches can be combined in the same PROC MI, allowing, for example, regression, propensity score, and discriminant analysis to be used in completing a single data set.
The propensity score approach fits a logistic regression model predicting the missingness indicator, then orders the observations by predicted probability of missingness. Next, the ordered observations are split into G equal-sized groups. Loosely, the imputed value is then chosen at random from among the observed values in the same one of the G groups as the missing observation (Lavori, Dawson & Shera 1995); G can be chosen by the user.
The logistic (or ordinal logistic) regression approach is the standard one proposed by Rubin (1987). Analogous to the logistic regression approach, the discriminant analysis approach creates a probability of each level of a categorical variable, then draws a random uniform variate and assigns an imputed level based on this. The distinction is that the probabilities are based on discriminant analysis, rather than logistic regression. The user has some control over details of the process used. The advantage of this approach is that nominally-valued variables can be imputed, rather than the ordinal values required for ordinal logistic regression.
The code to fit PROC/MI using an two stage imputation is displayed in Figure 20 (Appendix), with output in Figure 21 (Appendix). This approach exploits the particular patterns observed in the motivating example. Because so few observations were missing for TOTCHG (approximately 4% missing), 20 imputed datasets were created as a function of the fully observed variables (lines 1–5). Imputation was then carried out for the three remaining variables (T=ATYPE, S=SEASON, R=RACE). Lines 15–17 create a variable that describes the missingness pattern (111=fully observed, 112=race and type observed, season missing, etc.). Separate monotone imputations are carried out for each pattern, based on what is observed (lines 26–46) and these imputed values are merged into a single data (lines 48–52). The logistic regression model is fit for each imputed dataset (lines 56–61) and the results are consolidated using MIANALYZE (lines 72–76).
The three-stage approach used in SAS highlights the notion of using different software to impute, analyze, and combine methods. For example, if a desirable imputation package does not exist in one's preferred analysis package, one could impute using a stand-alone imputation package, save the resulting completed data sets, and import them for data analysis. The analyses could then be exported to a third package for combining the results. This approach is demonstrated using Amelia for imputation and SAS for analysis in Figure 6 (Appendix).
5.8 Missing data library for S-Plus
S-Plus version 7.0 includes a missing data library that extends S-Plus to support model-based missing data models using the methodology of Schafer (1997), by use of the EM algorithm (Dempster et al. 1977) and data augmentation (DA) algorithms (Tanner & Wong 1987). DA algorithms can be used to generate multiple imputations. The missing data library provides support for multivariate normal data ( impGauss), categorical data ( impLoglin) and conditional Gaussian models ( impCgm) for imputations involving both discrete and continuous variables.
Figure 22 (Appendix) displays the code to access the library and read in the data (lines 1–14), create imputed datasets (lines 18–22) and combine results (lines 23–29). Figure 23 (Appendix) displays the output from the S-Plus missing data library. Infinite values were created for four observations with missing TOTCHG, which led to non-convergence of the EM algorithm. These values were dropped in the imputations. This anomaly was reported to Insightful Technical Support, though resolution is unknown as of press time.
5.9 Other packages and routines
Other packages which provide support for imputation include Joseph Schafer's free software (macros for S-Plus and standalone windows package NORM), SOLAS, and SPSS. Schafer's software routines are an excellent companion to his book, but they do not support general purpose regression modeling and more modern implementations are available in S-Plus. SOLAS is designed specifically for the analysis of datasets with missing observations, and version 3.0 was reviewed previously (Horton & Lipsitz 1999). Because the current version of SOLAS (3.2) does not support estimation of logistic regression models, we did not fit models using the package. Support for missing data is included in the SPSS version 12.0 missing value library, reviewed by von Hippel (2004).
6 Missing data modeling in KID dataset
We now return to the analysis of the motivating example. Table 4 displays the results (in terms of log OR and SE) for several of the regression parameters for the complete case and incomplete data logistic regression models.
Table 4.
Results (in terms of log OR and SE) for selected regression parameters for a variety of incomplete data logistic regression models
| Package | WEEKEND | FEMALE | TOTCHG |
|---|---|---|---|
| complete case | −0.058 (0.026) | 0.089 (0.021) | −0.004 (0.0010) |
| Amelia II | −0.027 (0.020) | 0.103 (0.016) | −0.005 (0.0005) |
| Hmisc | −0.020 (0.020) | 0.099 (0.016) | −0.005 (0.0005) |
| ICE | −0.020 (0.020) | 0.099 (0.016) | −0.004 (0.0005) |
| IVEware | −0.021 (0.020) | 0.100 (0.016) | −0.004 (0.0005) |
| MICE | −0.021 (0.020) | 0.100 (0.016) | −0.004 (0.0005) |
| LogXact | −0.026 (0.020) | 0.105 (0.016) | −0.005 (0.0005) |
| SAS PROC MI | −0.036 (0.021) | 0.119 (0.017) | −0.003 (0.0006) |
| S-Plus | −0.018 (0.020) | 0.098 (0.016) | −0.004 (0.0005) |
Footnote: TOTCHG parameter represents change of $1,000
In general, the parameter estimates for the FEMALE and TOTCHG are quite similar for all missing data models relative to the complete case estimator. For the WEEKEND parameter, the 95% confidence interval for the complete case estimator would not include zero, which is not the case for the other models. The differing results for the WEEKEND parameter may indicate a selection bias due to discarding all the partially observed observations. The standard error estimates for the complete case estimator are as much as 30% larger than those of the missing data models.
7 Conclusion
It is critically important to address missing data, as it arises in almost all real world investigations. Accounting for incomplete observations is particularly important for observational analyses with many predictors. In our motivating example, no predictor was missing more than 16% of the time, yet 41% of observations had at least one missing value. Dropping all these observations and fitting a model to only the complete cases would be hugely inefficient and potentially biased.
In this paper, we have briefly described a series of principled methods that can be fit logistic regression models with incomplete data, reviewed their implementation in general purpose statistical software, and applied them to our motivating example.
We found that it is feasible to fit imputation models in practice, though there are some limitations, complications and shortcomings of current implementations. Additional time and effort is needed to specify models in addition to the primary focus of inference, further assumptions are required and compromises may sometimes be necessary. However, the value of this additional work is often justified by the potential increase in efficiency and decrease in bias.
While not a focus of our paper, sensitivity analyses are an important component of modeling when some data are missing, and should be routinely conducted. Such additional analyses require effort, but allow insight into the impact of missing data assumptions.
Reviews of methodologies used to account for missing values in practice indicate that for prognostic studies of cancer and research articles in the New England Journal of Medicine, use of principled approaches is relatively rare. Despite some of their limitations, the existence of these implementations should help to foster increased use of missing data methods in practice.
Supplementary Material
Footnotes
Helpful comments and assistance regarding details of the software implementations were provided by Frank Harrell Jr., Krista Kilmer, Gary King, Patrick Royston, Pralay Senchaudhuri, Stef van Buuren and Yang Yuan. We thank Suzanne Switzer for assistance with the review of missing data methods in the New England Journal of Medicine and James Carpenter, Amy Herring as well as Owen Thomas for useful comments. We are grateful for the support provided by NIMH grant R01-MH54693 and the Smith College Picker Program.
Contributor Information
Nicholas J. Horton, Department of Mathematics and Statistics Smith College, Northampton, MA.
Ken P. Kleinman, Department of Ambulatory Care and Prevention, Harvard Medical School and Harvard Pilgrim Health Care, Boston, MA
References
- Allison PD. Multiple imputation for missing data: a cautionary tale. Sociological Methods and Research. 2000;28:301–309. [Google Scholar]
- Allison PD. Missing data. SAGE University Papers; 2002. [Google Scholar]
- Allison PD. Imputation of categorical variables with PROC MI. [accessed July 30, 2006];2005 http://www2.sas.com/proceedings/sugi30/113-30.pdf.
- Barnard J, Meng XL. Applications of multiple imputation in medical studies: from AIDS to NHANES. Statistical Methods in Medical Research. 1999;8:17–36. doi: 10.1177/096228029900800103. [DOI] [PubMed] [Google Scholar]
- Bernaards CA, Belin TR, Schafer JL. Robustness of a multivariate normal approximation for imputation of incomplete binary data. Statistics in Medicine. doi: 10.1002/sim.2619. (In press) [DOI] [PubMed] [Google Scholar]
- Bishop YMM, Fienberg SE, Holland PW. Discrete multivariate analyses: Theory and practice. MIT Press; 1975. [Google Scholar]
- Burton A, Altman DG. Missing covariate data within cancer prognostic studies: a review of current reporting and proposed guidelines. British Journal of Cancer. 2004;91:4–8. doi: 10.1038/sj.bjc.6601907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J. Annotated bibliography on missing data. [accessed July 30, 2006];2006a http://www.lshtm.ac.uk/msu/missingdata/biblio.html.
- Carpenter J. Missing data example analysis. [accessed December 19, 2006];2006b http://www.lshtm.ac.uk/msu/missingdata/example.html.
- Carpenter J, Kenward M, Evans S, White I. Last observation carry-forward and last observation analysis. Statistics in Medicine. 2004;23:3241–3244. doi: 10.1002/sim.1891. [DOI] [PubMed] [Google Scholar]
- Carpenter J, Pocock S, Lamm CJ. Coping with missing data in clinical trials: a model based approach applied to asthma trials. Statistics in Medicine. 2002;21:1043–1066. doi: 10.1002/sim.1065. [DOI] [PubMed] [Google Scholar]
- Carpenter JR, Kenward MG, Vansteelandt S. A comparison of multiple imputation and doubly robust estimation for analyses with missing data. Journal of the Royal Statistical Society, Series A. 2006;169(3):571–584. [Google Scholar]
- Collins LM, Schafer JL, Kam CM. A comparison of inclusive and restrictive strategies in modern missing data procedures. Psychological Methods. 2001;6(4):330–351. [PubMed] [Google Scholar]
- Cook RJ, Zeng L, Yi GY. Marginal analysis of incomplete longitudinal binary data: a cautionary note on LOCF imputation. Biometrics. 2004;60:820–828. doi: 10.1111/j.0006-341X.2004.00234.x. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society, Series B. 1977;39(1):1–22. [Google Scholar]
- Diggle PJ, Heagerty P, Liang KY, Zeger SL. Analysis of Longitudinal Data. 2. Clarendon Press; 2002. [Google Scholar]
- Diggle P, Kenward MG. Informative drop-out in longitudinal data analysis. Applied Statistics. 1994;43:49–73. [Google Scholar]
- Glynn RJ, Laird NM, Rubin DB. Multiple imputation in mixture models for nonignorable nonresponse with follow-ups. Journal of the American Statistical Association. 1993;88:984–993. [Google Scholar]
- Greenland S, Finkle WD. A critical look at methods for handling missing covariates in epidemiologic regression analyses. American Journal of Epidemiology. 1995;142:1255–1264. doi: 10.1093/oxfordjournals.aje.a117592. [DOI] [PubMed] [Google Scholar]
- Harrell FE. Hmisc package. [accessed August 10, 2006];2006 http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/Hmisc.
- HCUP Kids' Inpatient Database (KID) Healthcare Cost and Utilization Project (HCUP) [accessed July 15, 2006];2000 doi: 10.1016/j.ijporl.2023.111767. URL: http://www.hcup-us.ahrq.gov/kidoverview.jsp. [DOI] [PMC free article] [PubMed]
- Honaker J, King G. What to do about missing values in time series cross-section data. [accessed December 17, 2006];2006 http://gking.harvard.edu/files/abs/pr-abs.shtml.
- Honaker J, King G, Blackwell M. Amelia software website. [accessed December 15, 2006];2006 http://gking.harvard.edu/amelia.
- Horton NJ, Laird NM. Maximum likelihood analysis of generalized linear models with missing covariates. Statistical Methods in Medical Research. 1999;8:37–50. doi: 10.1177/096228029900800104. [DOI] [PubMed] [Google Scholar]
- Horton NJ, Laird NM, Murphy JM, Monson RR, Sobol AM, Leighton AH. Multiple informants: Mortality associated with psychiatric disorders in the Stirling County Study. American Journal of Epidemiology. 2001;154(7):649–656. doi: 10.1093/aje/154.7.649. [DOI] [PubMed] [Google Scholar]
- Horton NJ, Lipsitz SR. Review of software to fit generalized estimating equation (GEE) regression models. The American Statistician. 1999;53:160–169. [Google Scholar]
- Horton NJ, Lipsitz SR. Multiple imputation in practice: comparison of software packages for regression models with missing variables. The American Statistician. 2001;55(3):244–254. [Google Scholar]
- Horton NJ, Lipsitz SR, Parzen M. A potential for bias when rounding in multiple imputation. The American Statistician. 2003;57(4):229–232. [Google Scholar]
- Horton NJ, Switzer SS. Statistical methods in the Journal (research letter) New England Journal of Medicine. 2005;353(18):1977–1979. doi: 10.1056/NEJM200511033531823. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG. Incomplete data in generalized linear models. Journal of the American Statistical Association. 1990;85(411):765–769. [Google Scholar]
- Ibrahim JG, Chen MH, Lipsitz SR. Monte Carlo EM for missing covariates in parametric regression models. Biometrics. 1999;55(2):591–596. doi: 10.1111/j.0006-341x.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Chen MH, Lipsitz SR, Herring AH. Missing-data methods for generalized linear models: A comparative review. Journal of the American Statistical Association. 2005;100(469):332–346. [Google Scholar]
- Imai K, King G, Lau O. Zelig software website. [accessed December 15, 2006];2006 http://gking.harvard.edu/zelig.
- Jansen I, Beunckens C, Molenberghs G, Verbeke G, Mallinckrodt C. Analyzing incomplete discrete longitudinal clinical trial data. Statistical Science. 2006;21(1):52–69. [Google Scholar]
- Jones MP. Indicator and stratification methods for missing explanatory variables in multiple linear regression. Journal of the American Statistical Association. 1996;91(433):222–230. [Google Scholar]
- Kenward MC, Carpenter J. Multiple imputation: current perspectives. Statistical Methods in Medical Research. doi: 10.1177/0962280206075304. (In press) [DOI] [PubMed] [Google Scholar]
- King G, Honaker J, Joseph A, Scheve K. Analyzing incomplete political science data: an alternative algorithm for multiple imputation. American Political Science Review. 2001;95:49–69. [Google Scholar]
- Laird NM. Missing data in longitudinal studies. Statistics in Medicine. 1988;7:305–315. doi: 10.1002/sim.4780070131. [DOI] [PubMed] [Google Scholar]
- Lavori PW, Dawson R, Shera D. A multiple imputation strategy for clinical trials with truncation of patient data. Statistics in Medicine. 1995;14:1913–1925. doi: 10.1002/sim.4780141707. [DOI] [PubMed] [Google Scholar]
- Lipsitz SR, Ibrahim JG. A conditional model for incomplete covariates in parametric regression models. Biometrika. 1996;83(4):916–922. [Google Scholar]
- Little RJA. Missing-data adjustments in large surveys. Journal of Business and Economic Statistics. 1988;6:287–296. [Google Scholar]
- Little RJA. Regression with missing X's: a review. Journal of the American Statistical Association. 1992;87(420):1227–1237. [Google Scholar]
- Little RJA. A class of pattern-mixture models for normal incomplete data. Biometrika. 1994;81:471–483. [Google Scholar]
- Little RJA, Rubin DB. Statistical analysis with missing data. John Wiley & Sons; New York: 1987. [Google Scholar]
- Little RJA, Rubin DB. Statistical analysis with missing data. 2. John Wiley & Sons; New York: 2002. [Google Scholar]
- Meng XL. Missing data: dial M for ??? Journal of the American Statistical Association. 2000;95(452):1325–1330. [Google Scholar]
- Moons KGM, Donders RA, Stijnen T, Harrell FE. Using the outcome for imputation of missing predictor values was preferred. Journal of Clinical Epidemiology. 2006;59(10):1092–1101. doi: 10.1016/j.jclinepi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Olkin I, Tate RF. Multivariate correlation models with mixed discrete and continuous variables. The Annals of Mathematical Statistics. 1961;32:448–465. [Google Scholar]
- Raghunathan TE. What do we do with missing data? some options for analysis of incomplete data. Annual Review of Public Health. 2004;25:99–117. doi: 10.1146/annurev.publhealth.25.102802.124410. [DOI] [PubMed] [Google Scholar]
- Raghunathan TE, Lepkowski JM, van Hoewyk J, Solenberger P. A multivariate technique for multiply imputing missing values using a sequence of regression models. Survey Methodology. 2001;27(1):85–95. [Google Scholar]
- Robins JM, Rotnitzky A, Zhao LP. Analysis of semiparametric regression models for repeated outcomes in the presence of missing data. Journal of the American Statistical Association. 1995;90:106–121. [Google Scholar]
- Royston P. Multiple imputation of missing values. Stata Technical Journal. 2005;5(4):527–536. [Google Scholar]
- Rubin DB. Inference and missing data. Biometrika. 1976;63:581–590. [Google Scholar]
- Rubin DB. Multiple Imputation for Nonresponse in Surveys. Wiley; 1987. [Google Scholar]
- Rubin DB. Multiple imputation after 18+ years. Journal of the American Statistical Association. 1996;91:473–489. [Google Scholar]
- Schafer JL. Analysis of incomplete multivariate data. Chapman & Hall; 1997. [Google Scholar]
- Schafer JL. Multiple imputation: a primer. Statistical Methods in Medical Research. 1999;8:3–15. doi: 10.1177/096228029900800102. [DOI] [PubMed] [Google Scholar]
- Smith GC, Wood AM, Pell JP, White IR, Crossley JA, Dobbie R. Second-trimester maternal serum levels of alpha-fetoprotein and the subsequent risk of sudden infant death syndrome. New England Journal of Medicine. 2004;351(10):978–986. doi: 10.1056/NEJMoa040963. [DOI] [PubMed] [Google Scholar]
- Tanner MA, Wong WH. The calculation of posterior distributions by data augmentation. Journal of the American Statistical Association. 1987;82:528–540. [Google Scholar]
- van Buuren S. Multiple imputation online. [accessed August 19, 2006];2006 http://www.multiple-imputation.com.
- van Buuren S. Creating multiple imputations in discrete and continuous data by fully conditional specification. Statistical Methods in Medical Research. doi: 10.1177/0962280206074463. (In press) [DOI] [PubMed] [Google Scholar]
- van Buuren S, Boshuizen HC, Knook DL. Multiple imputation of missing blood pressure covariates in survival analysis. Statistics in Medicine. 1999;18:681–694. doi: 10.1002/(sici)1097-0258(19990330)18:6<681::aid-sim71>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- van Buuren S, Brand JPL, Groothius-Oudshoorn CGM, Rubin DB. Fully conditional specification in multivariate imputation. Journal of Statistical Computation and Simulation. 2006;76(12):1049–1064. [Google Scholar]
- van de Beek D, de Gans J, Spanjaard L, Weisfelt M, Reitsma JB, Vermeulen M. Clinical features and prognostic factors in adults with bacterial meningitis. New England Journal of Medicine. 2004;351(18):1849–59. doi: 10.1056/NEJMoa040845. [DOI] [PubMed] [Google Scholar]
- von Hippel P. Biases in SPSS 12.0 missing value analysis. The American Statistician. 2004;58(2):160–164. [Google Scholar]
- Xie F, Paik MC. Generalized estimating equation model for binary outcomes with missing covariates. Biometrics. 1997;53:1458–1466. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


