Abstract
The use of fluorescent protein tags has had a huge impact on cell biological studies in virtually every experimental system. Incorporation of coding sequence for fluorescent proteins such as green fluorescent protein (GFP) into genes at their endogenous chromosomal position is especially useful for generating GFP-fusion proteins that provide accurate cellular and subcellular expression data. We tested modifications of a transposon-based protein trap screening procedure in Drosophila to optimize the rate of recovering useful protein traps and their analysis. Transposons carrying the GFP-coding sequence flanked by splice acceptor and donor sequences were mobilized, and new insertions that resulted in production of GFP were captured using an automated embryo sorter. Individual stocks were established, GFP expression was analyzed during oogenesis, and insertion sites were determined by sequencing genomic DNA flanking the insertions. The resulting collection includes lines with protein traps in which GFP was spliced into mRNAs and embedded within endogenous proteins or enhancer traps in which GFP expression depended on splicing into transposon-derived RNA. We report a total of 335 genes associated with protein or enhancer traps and a web-accessible database for viewing molecular information and expression data for these genes.
AS a model organism, perhaps the most important advantage of Drosophila is the extensive range of very sophisticated genetic tools available. Central among these are transposons, including P elements and piggyBac (PBac) elements, which can be used to create easily mapped insertion mutations and to analyze gene expression with a variety of reporter molecules. P elements are naturally occurring Drosophila transposable elements that were first modified to provide vectors for efficient DNA-mediated gene transfer in Drosophila (Rubin and Spradling 1982) and then to create collections of random, single-element insertions in the genome (Robertson et al. 1988; Cooley et al. 1989; Spradling et al. 1995, 1999; Bellen et al. 2004). Although very useful for mutagenesis, P elements also have limitations including preference for the 5′ region of genes (O'Hare and Rubin 1983; Tsubota et al. 1985), bias toward particular sequence motifs (O'Hare and Rubin 1983), and “hot spots” that have been hit at a high frequency (Spradling et al. 1999).
The more recent use of transposons based on the Lepidopteran PBac element has expanded the number of genes disrupted by single transposon insertions (Horn et al. 2003; Bellen et al. 2004; Bonin and Mann 2004; Thibault et al. 2004). First introduced into the Drosophila melanogaster germline by Handler and Harrell (1999), the PBac elements were shown to transpose and insert at TTAA sequences. Like the P element, PBac contains a single open reading frame encoding transposase and is bounded by short terminal inverted repeats. PBac elements have been demonstrated to insert into new genes that have not previously been hit using P-element techniques.
Engineered transposable elements have been productively employed to analyze gene expression by enhancer trapping, gene trapping, and, most recently, protein-trapping strategies. Enhancer trapping in Drosophila typically involves a transposon carrying a reporter gene, such as β-galactosidase or green fluorescent protein (GFP), linked to a weak basal promoter. Insertion in the genome near genes can result in activation of the promoter and expression of the reporter gene in a pattern similar to that of the endogenous gene (O'Kane and Gehring 1987; Wilson et al. 1989). This technology is useful in a variety of organisms including bacteria (Dunn and Handelsman 1999) and zebrafish (Balciunas et al. 2004).
From enhancer-trapping methods have evolved gene-trapping strategies, using transposons lacking a promoter. These elements rely on an endogenous gene's promoter and control region to report expression. First described in mouse embryonic stem cells (Gossler et al. 1989; Skarnes et al. 1992), these constructs usually use a splice acceptor site upstream of the reporter gene. Upon insertion into an intron, the splicing machinery incorporates, or “traps,” the element into a gene's normal transcription. Variations of the mammalian constructs contain the lac Z gene and were designed with a transcription termination sequence to increase the possibility of mutation (Skarnes et al. 1995; Cannon et al. 1999; Hirashima et al. 2004). GFP-tagged gene traps have also been generated in zebrafish (Kotani et al. 2006) and Drosophila (Bonin and Mann 2004).
In addition to a gene's transcriptional readout, its protein localization pattern can also be critical in deciphering gene function. Protein traps were developed to study protein localization without the need to make antibodies or use fixed tissue. Protein traps were first developed as “CD tagging” in Chlamydomonas and Drosophila (Jarvik et al. 1996) and later in mammalian cell lines as “mini-exon epitope tagging” (Smith 1997). Both of these methods use an open reading frame flanked by splice acceptor (SA) and donor (SD) sites. Upon insertion into an intron and subsequent transcription, a small peptide is incorporated into the endogenous protein. Since the tags are unique, specific antibodies can be used for functional studies. The concept was later adapted to use GFP as a tag, thus eliminating the use of antibodies, in Drosophila (Morin et al. 2001; Clyne et al. 2003). In these studies, GFP is used as a mobile artificial exon carried by a transposable P element. The GFP gene lacks initiation and stop codons and is flanked by SA and SD sites. When the intron phase matches the frame of the GFP exon, GFP is seamlessly inserted into an otherwise intact transcript and a GFP-expressing fusion protein is generated.
Like enhancer traps, protein traps provide a starting point for mutagenesis, information on gene expression, and easy identification of the trapped gene by sequencing the flanking genomic DNA. However, protein traps additionally provide information on the expression and subcellular localization of gene products. The use of GFP also allows a real-time dynamic study of the endogenous protein distribution in live tissue using flourescence microscopy. Other protein-trap vectors have recently been developed for use in systems such as mammalian cells (Sineshchekova et al. 2004).
Here, we report the results of a series of innovative protein-trap screens in D. melanogaster, which include the addition of a high-throughput embryo sorter, so that millions of animals were screened. We also describe the employment of both P and PBac transposable elements to generate a series of GFP-encoding protein traps and discuss the strategies, execution, results, and limitations of these approaches. This work was conducted in collaboration with our colleagues in the Chia laboratory in Singapore and the Spradling laboratory at the Carnegie Institution (see accompanying article by Buszczak et al. 2007, this issue).
MATERIALS AND METHODS
Fly stocks:
Flies were maintained under standard culturing conditions. The y,w/Y, P{hs:hid} virgining stock was obtained from Ruth Lehmann (New York University). P-element transposase stocks yw; Ki, P{Δ2-3}99B and yw; Sb, P{Δ2-3}99B were obtained from the Bloomington Drosophila Stock Center at Indiana University. Deficiency stocks for complementation tests of lethal lines were also obtained from the Bloomington Drosophila Stock Center at Indiana University (see supplemental Table 2 at http://www.genetics.org/supplemental/). P-element mutator lines w; P{PTT-GA}III-1b, w; P{PTT-GB}III-3b, and P{PTT-GC}III-4b were obtained from William Chia (Temasek Lifesciences Laboratory, Singapore). Lines in the “G” and “ZCL” collections (Morin et al. 2001) were obtained from William Chia.
P-element mutator lines:
P-element-based protein traps used in this work contain the enhanced green fluorescent protein (EGFP) gene encoded as a mobile artificial exon carried by a transposable P element (Morin et al. 2001). The SA and SD sequences from the Drosophila Myosin heavy chain II gene flank the EGFP coding sequence, which lacks translation initiation and termination codons. The construct also encodes a 6xHis epitope fused to the amino terminus of EGFP. The transposon is marked with the eye color selection gene, mini-white. Fly lines containing three independent constructs were used, each containing the EGFP gene flanked by splicing sequences for one of the three reading frames. Splicing of the EGFP exon in PTT-GA maintains the reading frame interrupted by a phase 1 intron, PTT-GB is compatible with phase 2 introns, and PTT-GC with phase 0 introns. To move the mutators onto the CyO balancer chromosome, the insertions in w; P{PTT-GB}III-3b and P{PTT-GC}III-4b were remobilized by crossing to P{Δ2-3}99B in the presence of CyO. Insertions of the P{PTT-GB} and the P{PTT-GC} mapping to CyO and not expressing EGFP were used for the YB and YC screens.
piggyBac mutator and transposase lines:
A PBac protein trap construct was made by inserting the 6xHisEGFP A-frame exon into pBac[D.m. w+] (Handler and Harrell 1999) and substituting the yellow gene for mini-white to create pBAC{HpaI-GFP; y+} (see accompanying article by Buszczak et al. 2007 for details). Transgenic flies were created using the phspBac helper plasmid (Handler and Harrell 1999), and the insertion on the X chromosome in pBAC{HpaI; y+}24.3 was remobilized to recover new insertions on the CyO balancer chromosome. An insertion on CyO that did not express EGFP was used in the YD screen.
A genomic source of piggyBac transposase was made by P-element-mediated transformation. The piggyBac transposase gene was excised from phspBac and joined with the ubiquitin promotor by cloning into the pCasper3-Up2-RX poly(A) P-element vector (Ward et al. 1998) to create P{w+, Ub-pBACtrans} (see Buszczak et al. 2007 for details).
Genetic crosses:
P screen:
The first strategy was a modification of that described in Morin et al. (2001). To accelerate the rate of recovering GFP-positive lines, we used a high-throughput automated Drosophila embryo sorter (COPAS Select; Union Biometrica). For the pilot screen, we used protein trap lines with the highest frequency of transposition (w; P{PTT-GA}III-1b, w; P{PTT-GB}III-3b, and P{PTT-GC}III-4b) (Morin et al. 2001). Virgin females carrying P{Δ2-3}99B were crossed to a mixed population of males carrying the GA, GB, or GC protein trap vector. All of the starting elements were on the third chromosome and did not express detectable GFP. Male progeny carrying both P{Δ2-3}99B and a protein trap were mated to y,w virgin females. A virgining stock, y,w/Y, P{hs:hid}, was used to facilitate collecting large numbers of virgins. Embryos and first and second instar larvae in standard bottles were heat-shocked in 37° sand pits for 2 hr on 2 consecutive days. Females were collected shortly after eclosion. Approximately 250 males and 750 virgins were placed in large culture containers (condos). The flies were allowed to acclimate in the condos for 2–3 days with daily feeding of wet yeast paste on grape juice agar plates. For embryo collection, females were allowed to lay embryos on a fresh plate for 12–20 hr. Embryos were collected and screened from the same parents for 2 consecutive days, and then the parents were discarded. Embryos were dechorionated by incubation in 50% bleach for 2 min and rinsed extensively in PBS. The embryos were suspended in 100–200 ml of PBS with 2% Tween-20 and transferred to the sample cup of the embryo sorter. Using multiple condos, between 50,000 and 200,000 embryos were collected for each sorting session. Embryos flowed from a continuously mixed sample cup to a preanalysis chamber, where the embryos are surrounded by a sheath solution (Union Biometrica) to produce a stabilized flow and focus the embryos in the center of the flow stream. Embryos pass through the flow cell where a red diode laser (670 nm) measures size and optical density, while a multiline argon laser excites the GFP fluorophore. Emission signals are measured and real-time analysis of the measured parameters is used to make sorting decisions. Only those embryos that are GFP positive are dispensed into the standard food vials. GFP-negative embryos are directed by a puff of air to a sample container where they are discarded. The embryo sorter examined ∼70,000 embryos/hr. After GFP-positive embryos completed development, single animals with eye pigment (w+) were mated to y,w flies to establish stocks. Only flies that did not inherit the P{Δ2-3}99B chromosome were used to establish stocks. Stocks produced in this pilot screen were labeled “P” followed by an isolation number.
YB and YC screens:
The screen strategy was modified by moving the starting elements to the CyO balancer chromosome. In addition, one reading frame was screened at a time. Crosses to generate embryos for screening by the COPAS Select embryo sorter were carried out as follows: (1) w/Y; CyO, P{w+, GB} (or CyO, P{w+, GC}) × w; Sb, P{Δ2-3}99B (or Ki, P{Δ2-3}99B) virgins and (2) w/Y; CyO, P{w+, GB} (or CyO, P{w+, GC}); Sb, P{Δ2-3}99B (or Ki, P{Δ2-3}) × y,w virgins.
Cross 1 was carried out in standard fly bottles, and cross 2 was done in condos as described above. Lines resulting from this screen were designated “YB” or “YC.” For the YC screen, only flies that did not inherit the P{Δ2-3} chromosome and the CyO chromosome were used to establish stocks. During the YB screen, we included flies carrying CyO for initial stocks. Only those lines in which CyO segregated from white+, indicating a new insertion, were kept.
YD screen:
To test the utility of PBac elements for protein trapping, the same EGFP artificial exon was mobilized in the context of a PBac transposon marked with yellow+. For this screen, the starting element encoded EGFP flanked by A-frame splicing signals compatible with the phase 1 introns. The crosses to produce embryos for screening were: (1) y,w/Y; CyO, PBac{y+, GA} × y,w; P{w+, Ub-pBACtrans} virgins and (2) y,w/Y; Cyo, PBac{y+, GA}; P{w+, Ub-pBACtrans} × y,w virgins.
Cross 1 was carried out in standard fly bottles, and cross 2 was done in condos as described above. Flies lacking the PBac transposase were identified by a white eye color. Only lines in which yellow+ segregated from CyO were kept. Lines resulting from this screen were designated “YD.”
Imaging ovarian expression patterns from protein traps:
Ovaries were dissected from females fed on wet yeast paste for ∼36 hr. Following dissection in IMADS buffer (Singleton and Woodruff 1994a), ovaries were fixed in 6:1 of heptane and devitellinizing buffer (6% formaldehyde, 16.7 mm potassium phosphate, pH 6.8, 75 mm potassium chloride, 25 mm sodium chloride, 3.3 mm magnesium chloride) for 10 min (Cooley et al. 1992). After washing in PBT (0.5% bovine serum albumin, 0.3% Triton X-100, PBS), egg chambers were separated and mounted in Antifade solution [30% glycerol, 2.5% DABCO (Sigma, St. Louis) in PBS]. Confocal images were acquired on a Zeiss LSM510 or LSM510 meta- (Yale Center for Cell and Molecular Imaging) or Bio-Rad (Hercules, CA) MRC600 laser-scanning microscope. Images were processed using Adobe Photoshop 7.0 and rotated to display with anterior on the left.
Analysis of DNA flanking protein trap insertions:
Genomic DNA flanking transposon insertions was amplified for DNA sequencing using inverse PCR (iPCR) or thermal asymmetric interlaced–PCR (TAIL–PCR). The procedure for iPCR was previously described (Liao et al. 2000; Bellen et al. 2004), and the detailed protocol is available on the website of the Drosophila Gene Disruption Project at http://flypush.imgen.bcm.tmc.edu/pscreen/. Amplification and sequencing of iPCR products was carried out either in the Cooley lab or in the Hoskins lab; amplification and sequencing of TAIL–PCR products was carried out in the Cooley lab. DNA sequencing at Yale was performed by the Keck Foundation Biotechnology Resource Laboratory.
iPCR:
For P-element insertions, genomic DNA was digested with the restriction enzyme HinP1I or Sau3A; 5′ flanks were amplified by iPCR with the primers Plac1 (CACCCAAGGCTCTGCTCCCACAAT) and Pwht1 (GTAACGCTAATCACTCCGAACAGGTCACA) at an annealing temperature of 60° and sequenced with 5.SUP.seq1 (TCCAGTCACAGCTTTGCAGC); 3′ flanks were amplified with EY.3.F (CCTTTCACTCGCACTTATTG) and EY.3.R (GTGAGACAGCGATATGATTGT) at an annealing temperature of 55° and sequenced with EY.3.F.
For PBac insertions, genomic DNA was digested with HaeIII; 5′ flanks were amplified with PLF (CTTGACCTTGCCACAGAGGACTATTAGAGG) and PLR (CAGTGACACTTACCGCATTGACAAGCACGC) at an annealing temperature of 65° and sequenced with PLR; 3′ flanks were amplified with PRF (CCTCGATATACAGACCGATAAAACACATGC) and PRR (AGTCAGTCAGAAACAACTTTGGCACATATC) at an annealing temperature of 55° and sequenced with PRF.
TAIL–PCR:
The procedure for TAIL–PCR was modified from Singer and Burke (2003). TAIL–PCR uses three nested primers complementary to the transposon and arbitrary degenerate (AD) primers that anneal to genomic locations. The annealing temperatures of the specific and AD primers differ, and PCRs with alternating cycles of high and low annealing temperatures allow amplification of specific products that include the junction of transposon and genomic DNA. AD primers were: AD1 (NGTCGASWGANAWGAA), AD2 (TGWGNAGSANCASAGA), AD3 (AGWGNAGWANCAWAGG), AD4 (STTGNTASTNCTNTGC), AD5 (NTCGASTWTSGWGTT), and AD6 (WGTGNAGWANCANAGA), where W equals A or T, S equals G or C, and N equals A or T or G or C. These were used singly or in various mixtures. Specific nested primers were designed for the 5′ and 3′ P-element ends, as well as for GFP in both directions. The 5′ P-element primers for TAIL1, TAIL2, and TAIL3 reactions were Plac1 (CACCCAAGGCTCTGCTCCCACAAT), Ep5-2 (TACTCCAGTCACAGCTTTGCAGCA), and Sp1 (ACACAACCTTTCCTCTCAACAA). The P-element 3′ primers for TAIL1, TAIL2, and TAIL3 reactions were Pry1out (ATTCAAACCCCACGGACATGCTAAGG), Pry4a (ACAATCATATCGCTGTCTCACTCAG), and Spep1a (CGACACTCAGAATACTATTCCTTTCAC). For GFP TAIL1, TAIL2, and TAIL3 reactions, the 5′ primer set consisted of Egfp-750R (CTACGGCAAGCTGACCCTGAA), Egfp-gsp1 (GAACTTGTGGCCGTTTACGTCGCC), and GgfpRT-A (GTCCAGCTCGACCAGGATGGGCAC), while the 3′ end was amplified with the primers gfpF1 (GGAGGACGGCAACATCCTGG), gfpF2 (CAACGTCTATATCATGGCCG), and gfpF3 (AGACCCCAACGAGAAGCGCG).
Genomic DNA was prepared by homogenizing 50 flies in 180 μl of PBS with a disposable microtube pestle and then isolated using the DNeasy Tissue kit membranes (QIAGEN, Valencia, CA) and eluted in 200 μl buffer AE (10 mm Tris–HCl, 0.5 mm EDTA, pH 9.0). For the TAIL1 PCR reaction, ∼600 ng genomic DNA were placed in a 20-μl reaction containing 2 μl 10× PCR reaction plus Mg buffer (Roche, Indianapolis), 0.4 μl 10 mm mixed dNTPs (Roche), 0.2 μl 5 units/μl Taq DNA polymerase (Roche), 1.5 μl 2 μm specific TAIL1 primer (listed above), 1.5 μl 40 μm mixed AD (or 40 μm AD3) primer (Singer and Burke 2003; Liu et al. 2005), and water. TAIL1 reactions were run in a Bio-Rad MyCycler thermal cycler programmed to settings decribed below. Reaction products were then diluted 1:50, 1 μl of which was used as template for 20-μl TAIL2 reactions (as TAIL1 reactions above, but substituting nested specific TAIL2 primer). For TAIL3 reactions, TAIL2 products were again diluted 1:50, and 1 μl was placed in a 20-μl reaction, as above, substituting nested specific TAIL3 primer. The thermal cycler settings for the three TAIL reactions were as follows: TAIL1, 94° for 3 min, 95° for 1 min, followed by 5 cycles of 94° for 1 min, 62° for 1 min, 72° for 2 min, 94° for 1 min, 25° for 3 min, ramping to 72° over 3 min, 72° for 2.5 min and 15 cycles of 94° for 30 sec, 65° for 1 min, 72° for 2.5 min, 94° for 30 sec, 65° for 1 min, 72° for 2.5 min, 94° for 30 sec, 44° for 1 min, 72° for 2.5 min, with an ensuing extension step of 72° for 10 min and cooling to 4°; TAIL2, 94° for 3 min, followed by 12 cycles of 94° for 30 sec, 64° for 1 min, 72° for 2.5 min, 94° for 30 sec, 64° for 1 min, 72° for 2.5 min, 94° for 30 sec, 44° for 1 min, 72° for 2.5 min, with a final extension step of 72° for 10 min and then cooling to 4°; TAIL3, 94° for 3 min, followed by 20 cycles of 94° for 1 min, 44° for 1 min, 72° for 2.5 min, with a final extension step of 72° for 10 min and then cooling to 4°.
TAIL3 PCR products were directly sequenced (unless multiple bands were visualized on an agarose gel in which case bands were extracted using a QIAquick gel extraction kit from QIAGEN) using the TAIL3-specific primer by the Keck Foundation Biotechnology Resource Laboratory at Yale.
Sequence analysis:
DNA sequences amplified by iPCR or TAIL–PCR were examined for the tranposon–genome junction by scanning for the proximal P or PBac ends. The distal junction made by iPCR was identified by text searching for the appropriate restriction site. All transposon and low-quality sequences were masked, and remaining sequences >20 bases were aligned to the D. melanogaster genome (Release 3.2) using BLASTN (Altschul et al. 1997). If sequences from both flanks aligned to the same position, the insertion site was assigned to the first base of the 8-bp target site duplication of P-element sequences or the 4-bp target site duplication of PBac insertions. If the flanking sequences aligned to different locations, the two were assumed to derive from separate insertions. If multiple alignments were present, the insertion was in repetitive DNA. When the proximal transposon–genome junction was not apparent in the flanking sequence, the insertion site was defined as the first base of the alignment to genomic DNA.
Analysis of GFP-containing transcripts in protein trap lines:
Ovarian RNA was isolated from lines that expressed EGFP during oogenesis. RT–PCR was used to determine the junction between EGFP coding sequence and the endogenous sequence in protein trap lines. Analysis of the 5′ and 3′ ends of GFP-containing transcripts was carried out by RACE.
RNA isolation:
Ovaries from 10 female flies fed on wet yeast paste were dissected in IMADS buffer (Singleton and Woodruff 1994a) using forceps cleaned in chloroform, then placed into 1.5-ml Eppendorf tubes containing 300 μl Trizol reagent (Invitrogen, San Diego) on ice, and homogenized within 30 min using disposable plastic micropestles (cleaned in chloroform). Homogenate was incubated at room temperature for 5 min, then 30 μl DEPC-treated water was added, and samples were vortexed vigorously before adding 60 μl chloroform and shaking tubes vigorously by hand. Samples were spun at 12,000 rpm for 10 min at 4°, and the aqueous layer was transferred to a new tube to which was added 0.8 vol isopropanol and then vortexed to mix and allowed to precipitate overnight at −20°. Samples were pelleted at 14,000 rpm for 30 min at 4°, supernatant was removed, and pellets were washed with 75% ethanol and allowed to dry 1–2 min at room temperature before dissolving in 50 μl DEPC-treated water. Following spectrophotometry, 10 μg of each sample were DNase treated in a 50-μl reaction, using the DNA Free kit (Ambion, Austin, TX).
3′ RACE:
For the first-strand reaction 1 μg of RNA was extended with 0.5 μm of AdP primer (GGCCACGCGTCGACTAGAACTTTTTTTTTTTTTTTTT) and 10 units of M-MuLV reverse transcriptase (Roche no. 11062603001) in a total of 20 μl at 42° for 50 min. The reaction was terminated with a 15-min incubation at 70°. The first PCR was performed with 2 μl of the first-strand reaction and 0.2 μm AdAmpP primer (GGCCACGCGTCGACTAGAAC) and 0.2 μm gfpF2 primer (CAACGTCTATATCATGGCCG) with an annealing temperature of 55°, an extension time of 3 min, and 30 cycles. Nested PCR was performed with 2 μl of the first PCR reaction and 0.2 μm AdAmpP primer (GGCCACGCGTCGACTAGAAC) and 0.2 μm gfpF3 primer (AGACCCCAACGAGAAGCGCG) with an annealing temperature of 55°, an extension time of 3 min, and 30 cycles. Individual bands were isolated (QIAGEN QIAquick gel extraction kit) and sequenced using the gfpF3 primer (W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University).
5′ RACE:
For the first-strand reaction 5 μg of RNA were extended with 0.5 μm of gfp R1 primer (GTCGTGCTGCTTCATGTGGTCG) and 10 units of M-MuLV reverse transcriptase (Roche no. 11062603001) in a total of 25 μl at 42° for 50 min. The reaction was terminated with a 15-min incubation at 70°. First-strand reactions were cleaned with the QIAquick PCR purification kit (QIAGEN) and eluted in 30 μl water. A terminal transferase reaction was then done with 10 μl of the first-strand reaction with 400 units terminal transferase (Roche no. 03333566001) and 0.2 μm dCTP. The first PCR was performed with 5 μl of dC-tailed cDNA, 0.2 μm AchP primer (GGCCACGCGTCGACTAGTTCGGGIIGGGIIGGGIIG), and 0.2 μm gfpR2 primer (GACACGCTGAACTTGTGGCCG) with an annealing temperature of 55°, an extension time of 3 min, and 30 cycles. Nested PCR was performed with 5 μl of the first PCR reaction diluted 1:100 and 0.2 μm AchAmpP primer (GGCCACGCGTCGACTAGTTCG) and 0.2 μm gfpR3 primer (AGCTCCTCGCCCTTGCTCACC) with an annealing temperature of 55°, an extension time of 3 min, and 30 cycles. Individual bands were isolated (QIAGEN QIAquick gel extraction kit) and sequenced using the gfpR3 primer (W. M. Keck Foundation Biotechnology Resource Laboratory, Yale University).
RT–PCR:
One-half microgram of DNase-treated total ovarian RNA was placed in a 50-μl reaction containing DEPC-treated water, 1 μl each of both 2 μm sense and 2 μm anti-sense primers, 25 μl reaction buffer and 1 μl RT/Taq enzyme from the SuperScript One-Step RT–PCR kit (Invitrogen). Reactions were processed in a GeneAmp PCR System 9700 thermal cycler (PE Applied Biosystems, Foster City, CA): RT at 50° 30 min, denaturation at 94° 2 min, followed by 40 cycles of 94° 30 sec, 55° 30 sec, 72° 1 min, with a final extension at 72° 10 min and cooling to 4°. PCR products were visualized on agarose gels and sequenced either directly or following gel extraction of bands using the QIAquick gel extraction kit (QIAGEN).
Western blots:
Ovaries were dissected in IMADS buffer (Singleton and Woodruff 1994b), homogenized in 10 μl sample buffer (10% glycerol, 2% SDS, 0.06 m Tris pH 6.8, 0.7 m β-mercaptoethanol) per ovary, and boiled for 5 min. Proteins were separated by SDS–PAGE electrophoresis and transferred to nitrocellulose. Polyclonal rabbit antisera against GFP (Torrey Pines TP401) was diluted 1:1000 in PBS with 5% powdered milk. Following overnight incubation at 4° in primary antibody, membranes were washed with PBS. Membranes were incubated for 1 hr with horseradish–peroxidase-conjugated secondary antibodies at 1:1000, and bands were detected using SuperSignal West Dura Extended Duration substrate (no. 34076; Pierce Chemical, Rockford, IL).
FlyTrap website:
Information on fly lines generated during this project and how to obtain them is available on our FlyTrap website (http://flytrap.med.yale.edu) (Kelso et al. 2004). This site is significantly enhanced with new searching tools and displays for insertion sites and images of GFP expression in tissues. FlyTrap was implemented using MySQL (version 5) (http://www.mysql.com). Our web server was implemented using Apache running on a Linux server. The web interface was built using PHP (http://www.php.net). The system also incorporates the open source gif-draw (GD) library V2 to generate the gene and insertion diagrams. Routine updates to the MySQL database are made from a FileMaker Pro (version 7) database using custom Perl scripts.
RESULTS
Automated embryo sorting:
The protein trap strategy used by Morin et al. (2001) involved screening first instar larvae with a dissecting microscope equipped to detect GFP fluorescence. Five thousand larvae could be examined per hour. We tested whether using an automated embryo sorter could accelerate the rate of screening for GFP+ animals. The embryo sorter was able to screen >70,000 embryos/hr and successfully retrieved 10 GFP+ embryos we placed among several thousand control embryos. The embryo sorter also performed well in screens for new protein trap insertions. We used the mutator lines described by Morin et al. (2001) and shifted from screening first instar larvae to screening embryos from 0–20 hr of development. Three P-element mutator lines, one for each reading frame (Morin et al. 2001) (see materials and methods), were mobilized by crossing to a source of P-element transposase (P{Δ2-3}99B). Males with both a mutator and P{Δ2-3}99B transposase were crossed to y,w virgins, and their progeny were screened using the embryo sorter. New insertions of a mutator element into a gene that result in production of GFP can be detected by monitoring GFP fluorescence. During the course of 3 months, we recovered >4000 GFP+ embryos from 2.5 million embryos (Table 1, P screen); however, low embryo survival (25%) reduced the number of lines that could be established to 1060. Lines were established from surviving fertile adults that did not inherit the P{Δ2-3}99B-bearing chromosome and designated P lines. We focused on examining ovaries for evidence of GFP-fusion protein expression and found that ∼75% of the lines had a GFP pattern during oogenesis. Thus, the embryo sorter can capture rare GFP+ animals and can improve the rate of screening for protein trap insertions.
TABLE 1.
Summary of screens
| Screen | No. embryos sorted | No. GFP positive | Survivors | Balanced lines | No. embryos sorted/line |
|---|---|---|---|---|---|
| P | 2,525,681 | 4,159 | 1,060 | 251 | 10,184 |
| YB | 3,786,000 | 2,120 | 1,713 | 217 | 17,609 |
| YC | 2,253,290 | 1,116 | 832 | 110 | 21,059 |
| YD | 10,140,000 | 3,279 | 1,929 | 433 | 23,098 |
| Totals | 18,704,971 | 10,674 | 5,534 | 1,049 |
Despite our success in producing new protein trap lines by screening embryos, having the starting positions for the mutator elements and the transposase on third chromosomes was a significant disadvantage. By selecting against P{Δ2-3}99B when choosing GFP+ animals, all initial lines set up contained the starting mutator elements. This complicated mapping and balancing of new insertions on the basis of following eye pigment produced by the white+ marker in the mutator. Reexamination of lines after balancing revealed many cases where the GFP+ pattern was lost. An additional complication was encountered when we attempted to determine the sequence of genomic DNA flanking protein trap insertions. New insertions on the third chromosome were accompanied by the starting element. Therefore, iPCR of third chromosome lines resulted in amplification of the starting elements as well as the protein trap insertions. The same problem occurred when sequencing flanking DNA from lines made by Morin et al. (2001). We tried to identify insertion sites in 132 G lines and 293 ZCL lines (Table 2) sent to us by William Chia. No informative sequence was recovered for 26 G lines and 107 ZCL lines. Most of the lines in which sequencing attempts failed mapped to the third chromosome: 14 of 26 G lines and 83 of 107 ZCL lines. Because of this, we focused on sequencing P lines that mapped to the second and X chromosomes and obtained informative sequence from 79 of 95 (Table 2). Therefore, the pilot screen showed that protein trapping with an automated embryo sorter was promising; however, the starting position of the mutator elements needed to be addressed.
TABLE 2.
Summary of sequencing insertion sites
| Screen | No. sequenced | Sequence recovered | Unique hit | Repetitivea | Double hitb |
|---|---|---|---|---|---|
| G | 132 | 110 | 105 | 4 | 1 |
| ZCL | 293 | 186 | 184 | 1 | 1 |
| P | 95 | 79 | 78 | 1 | 0 |
| YB | 206 | 180 | 176 | 3 | 1 |
| YC | 100 | 89 | 87 | 2 | 0 |
| YD | 407 | 404 | 388 | 13 | 3 |
| Total | 1233 | 1048 | 1018 | 24 | 6 |
The flanking sequence was from a transposable element that is located at many places in the genome.
The 5′ and 3′ flanking sequences mapped to different locations in the genome.
Screens with starting elements on CyO:
The mutator elements P{PTT-GB} and P{PTT-GC} were moved to the CyO balancer chromosome so that the chromosomes with starting elements could be followed with a dominant marker. We screened embryos that were the progeny of males carrying both CyO, P{PTT-G} and Ki (or Sb), P{Δ2-3}99B. This allowed us to select against both the starting mutator elements and the transposase source.
We screened >2.2 million embryos to obtain GFP+ embryos carrying insertions of P{PTT-GC} and nearly 3.8 million embryos for P{PTT-GB} (Table 1). These screens were done separately; therefore, the reading frame of the protein traps in the resulting lines was known. The date each GFP+ embryo was sorted was recorded and used to keep track of independent insertions. Lines made with P{PTT-GB} were designated with a YB prefix followed by an isolation number, and lines made with P{PTT-GC} were designated with YC. Survival of GFP+ embryos was improved compared to the pilot screen: 80% for YB embryos and 75% for YC embryos. Crosses to establish stocks and map insertions to chromosomes were set up from 1713 YB adults and 832 YC adults, resulting in 217 balanced YB lines and 110 YC lines (Table 1). There were several reasons the number of balanced lines was reduced compared with the initial number of adult survivors: death of the adults prior to mating, sterility of the adults, and discarding of stocks in which the new insert mapped to either the P{Δ2-3} chromosome or CyO. In addition, there were cases where no white+ progeny were produced; these were false positives collected by the sorter. We set the parameters for embryo collection close to the background level of GFP detection, reasoning that this would allow us to capture embryos with weak GFP expression. Overall for the YB and YC screens, one balanced line was produced for every ∼17,000 and ∼21,000 embryos sorted, respectively. This was lower than the rate for the P screen in which three mutator elements were mobilized simultaneously.
piggyBac screen:
The final screen we carried out used a PBac mutator that allows in-frame splicing of EGFP in the A reading frame. PBac has a simple AT-rich target sequence, TTAA (Cary et al. 1989), that allows PBac to insert into introns, which are relatively AT rich, more readily than P elements (Bonin and Mann 2004; Thibault et al. 2004). Therefore, we tested whether PBac-based protein trapping is more efficient than P-element screens.
Over 10 million embryos were sorted, resulting in 3279 embryos scored as GFP+ (Table 1). A total of 1929 animals survived to adulthood (59%) and were mated to y,w flies followed by crosses to map the insertions to chromosomal locations. A total of 433 balanced lines were produced and designated as YD lines. The overall production rate (1 line per 23,098 embryos screened) was similar to the rate for P-element mutators (Table 1).
Mapping and sequence analysis of insertion sites:
We mapped new insertions of protein trap mutators to chromosomal locations before analyzing expression patterns or sequencing insertion sites. We chose this strategy so that any expression patterns we found later were associated with mapped, stable inserts. Initial crosses of GFP+ animals occasionally produced progeny with two different (nonwhite) eye colors; independent lines were established and most cases were found to represent two independent insertions. Because we did not keep lines with insertions on the CyO chromosome (carrying the starting element) or the P{Δ2-3}-bearing chromosome, the available targets for new insertions were the X chromosome and one each of the major autosomes. Therefore, the expected distribution of inserts was 20% on the X, 40% on the second, and 40% on the third on the basis of the lengths of the euchromatic portions of these chromosomes. Genetic mapping of the lines made with P-element mutators (YB and YC) revealed marked deviations from this distribution. There were fewer insertions than expected on the second chromosome in both the YB and the YC lines (25%) (Table 3). The YC lines were skewed dramatically with 69% of insertions on the third chromosome and only 5% on the X chromosome. The distribution of insertions in the lines made with the PBac mutator (YD lines) was closest to the expected distribution (Table 3).
TABLE 3.
Chromosome distribution of insertions
| Chromosome | YB (% total) | YC (% total) | YD (% total) |
|---|---|---|---|
| X | 57 (26) | 6 (5) | 57 (13) |
| 2 | 53 (24) | 27 (25) | 170 (40) |
| 3 | 104 (48) | 74 (69) | 197 (46) |
| 4 | 1 (0.6) | 0 | 6 (1) |
To determine the insertion sites in the Y lines, the genomic DNA flanking these insertions was amplified, sequenced, and aligned with the D. melanogaster genome sequence (Release 3.2). We attempted to sequence most of the balanced lines—206 of 217 YB lines, 100 of 110 YC lines, and 407 of 433 YD lines—and informative sequence was obtained from >90% of the lines (Table 2). The vast majority (97%) of these lines contained a single insertion identified as a unique site by BLASTN, with the remaining lines having more than one insertion (double hit) or an insertion in repetitive DNA (Table 2). Thus, the modifications to the screening strategy were successful in reducing sequence failures due to the presence of starting elements.
Carrying out en masse matings to produce embryos for screening in the embryo sorter introduced the possibility of capturing premeiotic clusters of nonindependent transpositions. To produce a list of independent events among the YB, YC, and YD lines, we used genetic mapping data, the date the founding GFP+ animal was obtained, and the insertion site to identify duplicate events and remove them from further consideration. For the P lines, we used genetic mapping and insertion sites as criteria since the sort dates were not recorded for each line. For the G and ZCL lines we sequenced, we relied on the insertion site and the line designation to judge which lines had independent events. A total of 819 lines represent independent events (Table 4). In several instances more than one insertion was recovered per gene (Table 5); therefore, these insertions were associated with a total of 335 genes (see supplemental Table 1 at http://www.genetics.org/supplemental/ for complete list).
TABLE 4.
Trap type predicted by insertion site position relative to genes
| Screen | Independent insertions | % lethal | Protein | Protein?a | Enhancer | Enhancer opposite | No gene |
|---|---|---|---|---|---|---|---|
| G | 82 | ND | 55 | 0 | 9 | 9 | 9 |
| ZCL | 139 | ND | 93 | 2 | 12 | 22 | 10 |
| P | 60 | ND | 19 | 1 | 19 | 11 | 10 |
| YB | 159 | 33 | 36 | 1 | 54 | 46 | 22 |
| YC | 65 | 30 | 43 | 3 | 8 | 7 | 4 |
| YD | 314 | 47 | 49 | 10 | 152 | 30 | 73 |
| Total | 819 | 295 | 17 | 254 | 125 | 128 | |
| Genes | 335 | 115 | 4 | 136 | 82 | NA |
Examination of these insertions showed the potential for splicing the GFP exon to the first exon of a gene and maintaining an open reading frame. RNA analysis on these lines is needed to assign them to protein or enhancer trap designations.
TABLE 5.
Frequency distribution of targeted genes
| No. | P, YB, YC | G, ZCL | YD |
|---|---|---|---|
| 1 | 116 | 60 | 102 |
| 2 | 20 | 19 | 14 |
| 3 | 10 | 10 | 8 |
| 4 | 5 | 3 | 6 |
| 5 | 2 | 2 | 2 |
| 6 | 2 | 0 | 2 |
| 7 | 0 | 1 | 2 |
| 8 | 1 | 0 | 0 |
| 9 | 0 | 0 | 3 |
| 10 | 0 | 0 | 0 |
| 11 | 1 | 1 | 0 |
| 25 | 0 | 1 | 0 |
We balanced the insertions in the YB, YC, and YD lines and determined that 33% of the YB lines and 30% of the YC lines were homozygous lethal, while 47% of the PBac insertions in the YD lines were lethal (Table 4). We obtained viability information for 223 of the 255 genes associated with YB, YC, or YD insertions; lethal alleles were found in 104 of the genes, although 25 of these genes also had viable insertions. The remaining 119 genes had only viable insertions.
Insertions can create both protein and enhancer traps:
The gene for EGFP in the mutator lines lacks start and stop codons; therefore, expression of EGFP was expected to depend on insertion into an intron of a host gene and incorporation into the open reading frame of mature mRNAs by the splicing apparatus. We analyzed the position of insertions relative to nearby genes to predict whether they were within coding regions and found evidence not only for the expected protein traps, but also for enhancer traps. We included G and ZCL lines (Morin et al. 2001) that we sequenced, the P lines from the pilot screen, and the Y lines in this analysis.
Protein traps:
A total of 295 of the insertions, representing 115 genes (Table 4), were in introns between annotated protein-coding exons. These insertions were positioned to function as protein traps. Analysis of GFP expression in ovaries confirmed that the EGFP-fusion proteins had specific subcellular localization patterns. We obtained protein traps in both known and predicted genes that had specific expression patterns in the ovary. For example, lines with insertions in I'm not dead yet (Indy), which encodes a plasma membrane cation transporter (Knauf et al. 2002), localized specifically to follicle cell membranes (Figure 1A). Protein traps in Rtnl1, an endoplasmic reticulum (ER) protein (Wakefield and Tear 2006), showed EGFP localization to the ER in nurse cells and oocytes (Figure 1B). Insertions in calmodulin (Cam) produced GFP fluorescence localized primarily to cell membranes and less so to nuclei (Figure 1C). Finally, insertions in trailer hitch (Wilhelm et al. 2005) expressed GFP specifically in germline cytoplasm with enrichment in the oocyte (Figure 1D). Insertions in uncharacterized annotated genes also had specific subcellular localizations. For example, insertions in CG6416 expressed GFP in the ovarian muscle sheath (Figure 1E), and insertions in CG15926 resulted in GFP localization to follicle cell membranes (Figure 1F).
Figure 1.—
Protein trap expression patterns. (A) Expression of GFP inserted in I'm not dead yet (Indy) localizes to follicle cell membranes (line YC0017). (B) Expression of GFP inserted in Rtnl1 localizes to endoplasmic reticulum (line G00071). (C) Expression of GFP inserted in Calmodulin (Cam) localizes to nurse cell and oocyte membranes and also to nurse cell nuclei (line P00695). (D) Expression of GFP inserted in trailer hitch (tral) localizes germline cytoplasm and is enriched in the oocyte (line G01240). (E) Expression of GFP inserted in CG6416 localizes to muscle sheath (line ZCL0663). (F) Expression of GFP inserted in CG15926 localizes to follicle cell membranes (line G00035). Bar, 50 μm.
We carried out RT–PCR on a subset of the protein trap lines to provide molecular evidence that the GFP-coding exon was incorporated into mature mRNAs. We confirmed that 34 lines do fuse the GFP gene in frame with coding sequence from their associated gene. Thus, any lines in which insertions were within the coding sequence of a gene were designated as protein traps (Table 4). The fluorescence data coupled with the RT–PCR results provide further evidence that the protein traps are useful reporters of endogenous protein expression and localization.
Enhancer traps:
Unexpectedly, we found that many lines had insertions not predicted to be protein traps, suggesting that GFP was produced by other mechanisms. A total of 254 lines (Table 4, Enhancer) had insertions within 500 nucleotides of the 5′ end of a gene and with the GFP gene in the same 5′–3′ orientation, but upstream of the annotated protein-coding sequence. Thus, production of GFP in these lines depended on the availability of an upstream methionine codon. The GFP fluorescence was nuclear (Figure 2, A, C, and G) in 234 of 254 of these lines. Western blot analysis of ovary extracts probed with antibodies to GFP showed most bands to be similar in size, ∼40 kDa (Figure 2I, solid arrowhead), which is ∼14 kDa larger than GFP alone. We found two additional types of insertions: those that were inserted within an annotated gene but in the opposite orientation (Table 4, enhancer opposite) and those that were inserted >500 nucleotides from any annotated genes (Table 4, no gene). These lines also had almost exclusively nuclear GFP fluorescence patterns (Figure 2, B and D–F) and had GFP bands on Western blots of similar sizes to those seen in enhancer lines (Figure 2I, solid arrowhead). The highest frequency of enhancer, enhancer opposite, and no gene traps was in the YB and YD lines: 122 of 159 YB lines and 255 of 314 YD lines (Table 4).
Figure 2.—
Expression and mRNA analyses of enhancer trap and no gene lines. Ovarian expression patterns of GFP in enhancer trap lines (A, C, and G–H) and no gene lines (B and D–F) are shown. (A) GFP localizes to germline nuclei in enhancer trap line YD0184. (B) GFP localizes to follicle cell nuclei in no gene line YD0570. (C) GFP localizes to follicle cell nuclei in enhancer trap line YB0172. (D) GFP localizes to nuclei in no gene line ZCL2825. (E) GFP localizes to follicle cell nuclei in no gene line ZCL2860. (F) GFP localizes to nuclei in no gene line YB0147. (G) GFP localizes to nuclei and is concentrated in the germinal vesicle and puncta in the nurse cell nuclei (arrow) in enhancer trap line YB0011. (H) GFP localizes to the cytoplasm in enhancer trap line ZCL3170. (I) Western analysis with anti-GFP antibody of enhancer trap (Enh) and no gene (NG) lines. w1118 is the control showing several background bands recognized by the antibody. Circled numbers indicate the type of 3′ end seen in P-element insertions (see J). Lines YD0184, YD0570, YB0172, ZCL2825, ZCL2860, and YB0147 show protein products that run at ∼40 kDa (solid arrowhead). Line YB0011 shows a protein product at ∼110 kDa and ZCL3170 shows a protein at ∼75 kDa (open arrowheads). (J) Splicing schematic of P-element lines. In all lines examined, exon 0 of the P-element transposase gene (dark blue), which contains the methionine codon, splices in frame with the GFP. This splice completely removes the white gene that is in the opposite orientation to both the P element and the GFP gene. There are three distinct types of mRNA 3′ of GFP: (1) Readthrough of the splice donor (pink), which adds one amino acid followed by a stop codon and is followed by the P-element poly(A) addition signal 197 nucleotides downstream; (2) splicing from GFP to an exon with a noncanonical SA in the genome adding between 1 and 19 amino acids and a poly(A) addition signal; and (3) splicing from GFP to an annotated exon that contains the start codon of a known gene that is in frame with GFP with no intervening stop codon in the linker sequence. (K) Splicing schematic of PBac lines. All lines investigated showed the exact same splicing pattern. The most 5′ sequence is an annotated noncoding exon (red) that splices into two cryptic exons (light purple) in the 5′ PBac end (dark purple). The second PBac “exon” contains the start codon (Met) and the first 91 codons of the PBac transposase. Splicing from the second PBac exon to the GFP exon maintains the open reading frame. Splicing 3′ of GFP is to a cryptic exon that lies in the yellow gene. The cryptic exon is upstream of, and in the opposite orientation to, the yellow coding sequence. Six additional codons are in the cryptic exon, followed by a poly(A) addition signal.
To determine how GFP is expressed from these non-Protein trap lines, we produced and sequenced cDNAs of the GFP-containing mRNAs from a subset of the lines. cDNAs from both the P-element insertions in the YB, G, and ZCL lines and the PBac insertions in the YD lines included sequence from the transposon vectors (Figure 2, J and K). In all of the P-element lines analyzed, the GFP-containing cDNAs began with exon 0 of the P-element transposase gene, which is present in the 5′ end of the P-element vectors. Splicing of P-element exon 0 to the GFP exon fuses an in-frame AUG start codon and the first 97 amino acid codons of P-element transposase to the amino terminus of the GFP exon. The fusion of P-element transposase exon to the GFP exon is predicted to result in 39 kDa containing the transposase nuclear localization signal. All of the G and ZCL lines analyzed contained a GFP with a phase 2 splice acceptor sequence, the same phase that is in the YB lines. Therefore, the 5′ P end provided the transcriptional start site and translation initiation codon from P-element transposase. Insertions near strong transcriptional control sequences were apparently sufficient to activate transcription and produce GFP fused to an N-terminal fragment of P-element transposase.
We found three different types of cDNA sequence downstream of the GFP exon in the P-element lines. In five lines (three enhancers and two no genes), the transcript was unspliced and continued through the splice donor sequence (Figure 2J, circled 1). This introduced one additional amino acid codon followed by a stop codon. A polyadenylation signal was present 196 bases from the end of the GFP coding sequence within the 3′ P end. Other lines had evidence of splicing to downstream exons. In nine lines designated “no gene,” the GFP exon spliced into an unannotated genomic exon with partial canonical SA sites and poly(A) signal. These added between 1 and 19 codons to the end of the GFP. The lack of significant coding sequence and the noncanonical SA sequences indicate that these are most likely cryptic exons and not unannotated genes (Figure 2J, circled 2). None of these 3′ splice variants add significantly to the size of the predicted GFP-containing protein produced.
The third type of cDNA sequence found downstream of GFP in these lines was the first coding exon of an annotated gene where the reading frame was in frame with that gene (Figure 2J, circled 3). This was the case for YB0011, which is 5′ of the D1 chromosomal protein (D1) gene, and ZCL3170, which is 5′ of string of pearls (sop). The use of the Met codon from exon 0 of the P-element transposase gene allows for what is essentially an N-terminal tag to the native protein with a short linker sequence between. In support of this, the proteins seen on Western blots were much larger than those from lines that appear to be enhancer traps (Figure 2I, open arrowheads). In addition, the expresson pattern in egg chambers for these two lines was consistent with the type of proteins encoded by the genes. D1∷GFP is present in nuclei with prominent dots in nurse cell nuclei (Figure 2G). Sop is a ribosomal protein, and Sop∷GFP is located in the cytoplasm of both germline and follicle cells (Figure 2H). Although we originally designated these lines as enhancer traps because the insertion sites were 5′ of genes, the cDNA sequence, Western blot data, and expression patterns supported reclassifying these as protein traps.
RNA analysis of PBac enhancer traps revealed a splicing pattern in each of five lines examined that results in an identical predicted GFP-containing protein. In all cases, splicing 3′ of GFP was to the yellow genomic region in the vector (Figure 2K). This “exon” is not part of the yellow gene but lies in the upstream genomic region and on the strand to the yellow coding region. Like the downstream cryptic exons seen in the P-element lines, the cryptic exon in yellow contained a noncanonical SA site and poly(A) addition signal, but resulted only in six amino acid codons before a stop codon. Additionally, all lines analyzed showed 5′ splicing to sequences from the 5′ PBac end (Figure 2K). Two segments of the 5′ PBac end were incorporated into cDNAs, neither of which is a bona fide exon. The second segment contained the start codon of PBac transposase, which normally reads through the entire length of the 5′ PBac sequence. However, in these lines splicing results in the first 91 amino acids being fused to GFP, resulting in a predicted protein of 39 kDa. Unlike the P-transposase∷GFP fusion protein, there is no predicted nuclear localization sequence in the PBac-transposase∷GFP fusion protein.
These results suggest that the large number of non-protein traps was isolated due to splicing in frame to sequences in the 5′ P-element and PBac ends, thus contributing an upstream start codon to the GFP exon so that it can act as an enhancer trap. These lines are useful indicators of which cells express the trapped gene, but, unlike the protein traps, they do not provide information on endogenous protein localization.
Lethality as a function of trap type:
Once the trap types were known, we could determine the fraction of each that caused lethality among the YB, YC, and YD balanced lines (Table 6). Of the 81 genes in these lines with Protein trap insertions, we determined the homozygous phenotype for 70 and found that 36% had only lethal inserts. In contrast, for the enhancer trap insertions, 53% of the genes investigated had only lethal insertions. This difference may be because the enhancer trap insertions are near the transcription start sites and more likely to disrupt expression of the genes. The enhancer opposite type of insertion had the lowest rate of lethality; 29% of the examined genes had only lethal insertions. The lower lethality of enhancer opposite traps compared to enhancer traps is probably because enhancer opposites were not restricted to the vicinity of transcription start sites, but were distributed along the length of the genes.
TABLE 6.
Lethality as a function of trap type
| Trap typea | No. of genes | Phenotype determined | No. lethal | % lethal |
|---|---|---|---|---|
| Protein | 81 | 70 | 25 | 36 |
| Enhancer | 121 | 104 | 55 | 53 |
| Enhancer opposite | 70 | 65 | 19 | 29 |
The data are only for the lines designated YB, YC, and YD.
For eight genes, we recovered both lethal and viable independent protein trap insertion lines. The expression of GFP in both types of line was identical, indicating that the proteins themselves appeared to localize correctly. This raises uncertainty about whether lethality is caused by the insertions themselves or another lesion on the chromosome. Although we did not find evidence for multiple insertions, other lesions could be present. In support of this, we tested two genes with both viable and lethal insertions by crossing to deficiencies that uncover the genes (see supplemental Table 2 at http://www.genetics.org/supplemental/). In both cases the lethal insertions were viable over a deficiency. Therefore, we scored all eight genes as viable for the purposes of calculating overall lethality.
To test whether transposon insertions caused the observed lethality, we focused on the protein traps that were homozygous lethal and determined whether the lethality was complemented by deficiencies that delete the insertion sites. Deficiencies were available from the Bloomington Stock Center for lethal protein trap insertions in 17 of the 25 genes (Table 6). Of these, 10 were lethal over a deficiency, one was male lethal over a deficiency, and six were viable (see supplemental Table 2 at http://www.genetics.org/supplemental/). These data suggest that the lethality in ∼35% of the protein trap lines was not due to the insertion we mapped. Therefore, lethal lines must be tested for secondary lesions before continuing with analysis of the genes.
Protein traps can reveal isoform-specific expression patterns:
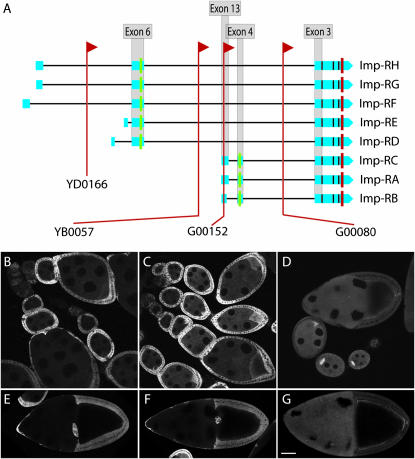
Multiple protein trap insertions in a gene of interest can be valuable for determining expression patterns of protein isoforms encoded by different transcripts. One example we discovered through our protein-trapping screen is the IGF-II mRNA-binding protein (Imp) gene for which we collected a total of 16 independent lines, incorporating both P and PBac elements. Imp encodes eight annotated protein isoforms by alternative splicing. The insertion sites were arrayed widely across the span of the gene within introns (Figure 3A). The insertions in the YD lines (PBac elements) were 5′ of all annotated start codons (Figure 3A) and had very low GFP expression during oogenesis. For the P-element insertions, we found several distinguishable expression patterns, three of which are shown in Figure 3. The G00080 line had the most 3′ insertion sites, and expression of GFP was in the cytoplasm of both the germline and the somatic follicle cells throughout oogenesis (Figure 3, D and G). GFP expression in G00080 showed a dramatic oocyte enrichment beginning in region 2 of the germarium (Figure 3D). In stage 10 egg chambers, GFP fluorescence was enriched subcortically in oocytes with the highest level of protein at the posterior pole (Figure 3G). The insertion in G00152 represents a group of insertions within the first exon of Imp-RA, Imp-RB, and Imp-RC, but still within an intron of the remaining transcripts. Expression was low within the germline, but strong in follicle cells throughout oogenesis (Figure 3, C and F). There was a slight enrichment of GFP in the oocytes in G00152 egg chambers (Figure 3C). YB0057 represents one of the insertions located upstream of Imp transcripts Imp-RA, Imp-RB, and Imp-RC (Figure 3A). GFP expression in YB0057 was highest in somatic follicle cells (Figure 3, B and E), and, in contrast to G00152 (Figure 3C), expression was reduced during stages 7–9 (Figure 3B).
Figure 3.—
Multiple insertions in Imp reveal isoform-specific expression patterns. (A) The IGF-II mRNA-binding protein (Imp) gene in FlyBase (FBgn0030235) has eight annotated transcripts, designated Imp-RA through Imp-RH. Insertion sites of the GFP protein traps in YD0166, YB0057, G00152, and G00080 are marked with red lines. All lines produced mRNAs with splicing of the GFP exon to Imp exon 3, which is contained in all the mature transcripts. Splicing 5′ of the GFP exon in G00080 incorporated exons 13 and 4 of the Imp-RC transcript. Splicing 5′ of the GFP exon in G00152 and YB0057 incorporated exon 6, but none of the transcript-specific exons upstream of exon 6 were detected in our RT–PCR analysis. (B–F) Confocal micrographs illustrating the expression patterns for the Imp insert lines YB0057 (B and E), G00152 (C and F), and G00080 (D and G). YB0057 had strong GFP expression in the somatic follicle cells, especially during stages 5–7, and less expression in the germline, with very little oocyte enrichment. G00152 expression was strong in follicle cells throughout oogenesis and showed some oocyte enrichment. G00080 expressed GFP in the germline throughout oogenesis, with strong oocyte enrichment and cortical and posterior localization in late oocytes, while follicle cell expression was reduced in comparison with the other insertions.
We carried out RT–PCR experiments to confirm that the GFP exon was incorporated into Imp transcripts. The current annotation of Imp (Grumbling and Strelets 2006) shows eight transcripts. We identified fusions between GFP coding sequence and Imp exon 3 (Figure 3A) in all 16 lines. The open reading frame was maintained in all cases such that GFP could be embedded in Imp proteins. 5′ RT–PCR was done on representative lines for each expression class. We found evidence of a transcript in the G00080 line containing exons 4 and 13, consistent with incorporation of GFP-coding sequence into the Imp-RC transcript (Figure 3A). None of the other lines analyzed had transcripts including exon 4. This suggests that the Imp-RC transcript is responsible for producing Imp protein that is highly enriched in the oocyte in this line. We tried to determine which Imp transcripts incorporated the GFP exon in G00152 and YB0057, which represent the other expression classes. Both these lines produced a fusion of GFP with exon 6, which is incorporated into the Imp-RD through Imp-RH transcripts. We used primers specific to the 5′-most exons of Imp-RD through Imp-RH, but were unable to find evidence of incorporation of the GFP exon into any of these transcripts. This suggests that additional Imp mRNAs with expression in follicle cells remain to be elucidated.
Comparing P-element and piggyBac protein traps:
We acquired confocal micrographs of adult ovarian tissue from a total of 431 P-element lines and 96 PBac lines and noted that the PBac lines generally appeared to have lower expression levels than the P-element lines. This was the case both for protein traps and for enhancer traps. Ninety-four percent of the P-element lines had expression in adult ovaries, 15% of which we classified as “faint.” In contrast, 70% of the PBac lines had expression during oogenesis, and 60% of these were faint.
To investigate this in more detail, we compared expression levels produced by PBac and P-element protein traps in several genes for which we recovered insertions of both elements (Figure 4). Since we had only PBac lines with the “A” reading frame, we compared these to P elements also carrying the A frame. These were available in the G and ZCL lines that we sequenced. The PBac insertion in all four genes we examined in detail produced weaker GFP fluorescence than a P-element insertion within the same intron. Although weaker, the localization of GFP was the same. Fasciclin3∷GFP was expressed highly in the follicle cells of the germarium and later resolved to specific expression in the polar follicle cells (Figure 4, A and B). This matches published Fasciclin3 antibody localization (Lee et al. 1997; Han et al. 2000). Sgg∷GFP was present in the cytoplasm of both nurse cells and follicle cells, with some enrichment on plasma membranes and nurse cell ring canals (Figure 4C). This expression is similar to that reported in Bobinnec et al. (2006). Expression from the PBac insertion in sgg was similar to the P-element insertion, but less intense (Figure 4D). The Oda gene (also called gutfeeling) encodes Ornithine decarboxylase antizyme. The P element and PBac protein trap insertions were both within the first intron of Oda, such that GFP tags the Oda-PA isoform. In egg chambers Oda∷GFP was present in both the cytoplasm and the nuclei of nurse cells and follicle cells (Figure 4E). Although localization of Oda has not been reported previously, the Oda gene has been shown to regulate Sex-lethal in the germline (Vied et al. 2003) so its presence in egg chambers was expected. Again, the level of expression produced in the PBac line was lower than that in the P-element line (Figure 4F). Finally, we compared expression of GFP∷Pdi from P-element and PBac protein traps. Protein disulfide isomerase (Pdi) is present in the lumen of the ER, and GFP∷Pdi expressed from P-element protein traps have been used to reveal ER dynamics during oogenesis (Bobinnec et al. 2003). We found the same pattern of expression of GFP∷Pdi from PBac protein traps in Pdi, although the level of expression was again lower compared to that from the P-element protein traps (Figure 4, G and H).
Figure 4.—
Comparison of GFP expression from P-element and PBac Protein traps. Confocal projection images of P-element and piggyBac protein traps inserted in the same gene are shown. (A and B) Protein traps inserted in Fasciclin3 (Fas3). Prominent expression of GFP is seen in follicle cell subsets including the cells that envelop germline cysts in the germarium (brackets), the stalk cells (arrowheads), and the polar cells (arrows). GFP fluorescence was much weaker in the pBac trap, as B was imaged with increased gain and laser power relative to the P-element trap in A (inset in B shows part of the pBac ovariole imaged under the same conditions as A). (C and D) Protein traps in shaggy (sgg) captured using identical imaging conditions to allow for an approximate comparison of GFP-fusion protein levels. Uniform cytoplasmic fluorescence is seen along with enrichment on plasma membranes. (E and F) Protein traps in Ornithine decarboxylase antizyme (Oda). The same pattern of nuclear enrichment is seen with both types of traps, although gain and laser power had to be increased when imaging the PBac trap. (G and H) Protein traps in Protein disulfide isomerase (Pdi). Pdi exhibits a characteristic ER subcellular localization pattern: nuclear envelope enrichment and a fenestrated cytoplasmic distribution. Again, GFP fluorescence in the PBac line was significantly weaker, requiring increased PMT gain relative to the P-element line. One notable difference between the two trap types in Pdi was the high level of GFP∷Pdi present in follicle cells relative to germ cells in the P-element insertion; this difference was not apparent in the PBac trap. Bars, 100 μm.
FlyTrap website:
Information on lines generated by this project is available in our web-accessible database FlyTrap (http://flytrap.med.yale.edu). The site also includes information on the G and ZCL lines made by Morin et al. (2001) and lines made in the Spradling lab (see accompanying article by Buszczak et al. 2007, this issue). FlyTrap houses information on insertion sites, associated genes, and images of expression patterns. It supports queries by gene name, line identifier, and gene ontology (GO) terms. In addition, fly stocks may be ordered through the website.
DISCUSSION
Since the discovery in 1994 that the GFP protein from the jellyfish Aequorea victoria can be expressed in other organisms (Chalfie et al. 1994), it has become widely used to tag proteins of interest in virtually every experimental system. GFP-tagged proteins are uniquely useful for examining the localization of the tagged protein in cells. Not only does GFP tagging provide an alternative to using antibodies for determining protein expression and localization patterns, but also the ability of GFP to fluoresce in living cells provides the opportunity to follow dynamic changes in protein localization during cellular events or in mutant backgrounds. Therefore, efforts to annotate the function of both characterized and uncharacterized proteins can be greatly aided by the availability of libraries of GFP-tagged proteins, as has been demonstrated in budding yeast (Huh et al. 2003).
To help reach the goal of producing GFP fusions to the majority of endogenous Drosophila proteins, we have explored transposon-mediated approaches for introducing an artificial GFP exon into genes (Morin et al. 2001). Because productive protein traps require insertion into introns, this approach has the advantage of introducing tags into genes at their normal chromosomal location with potentially minimal damage to promoters. The resulting protein expression patterns are controlled by the normal transcriptional control sequences and reflect endogenous expression. However, since the GFP-coding sequence must be in the same orientation as the host gene, and the reading frame of the GFP gene must match the intron phase, insertion events that produce bona fide protein traps are rare. Therefore, we used a high-throughput embryo sorter to accelerate the rate of recovering protein traps. We screened >18 million embryos to obtain a pool of >5000 animals carrying potential new protein traps. Analysis of the resulting insertions revealed that we recovered not only the expected protein traps, but also a variety of enhancer traps that result from unexpected splicing patterns. Our work both validated the use of transposons to produce protein traps and provided key insights into the limitations of current configurations of protein trap constructs.
Trap types:
Extensive sequence and expression analysis showed that, as expected, we identified protein trap insertions within introns that interrupt coding regions. A total of 295 fly lines were associated with 115 genes, including both characterized and uncharacterized genes. By RNA analysis, we confirmed that the GFP coding sequence is fused to predicted exons in 34 lines examined. This strongly suggests that all the protein traps are behaving as expected. Even though we screened for GFP-fusion proteins during embryogenesis, >70% of the lines were also expressed during oogenesis where they displayed a range of expression patterns revealing both cell-specific expression and subcellular localization. For example, Indy and CG15926 were expressed only in follicle cells, Fas3 was in polar follicle cells, while Rtnl and Trailer hitch were almost exclusively expressed in the germline. We observed many examples of uniform cytoplasmic localization, nuclear localization, association with plasma membranes, endoplasmic reticulum, and muscle fibers. Furthermore, in cases where we could compare, the majority of the protein trap expression patterns were similar to antibody staining patterns. Therefore, these lines will be useful for examining expression patterns in other cell types.
Although this screen was explicitly designed for the purpose of engineering protein traps, we in fact identified enhancer traps in more genes than we did for protein traps (136 enhancer traps vs. 115 protein traps). The enhancer traps were almost exclusively in the P-element lines that contained the B reading frame and the PBac lines that contained the A reading frame. RNA analysis indicated that this was due to splicing of sequences within the 5′ portion of the vector to provide a methionine start codon in frame with the coding region of the GFP. In the case of P-element enhancer traps, it appears that transcription can initiate in the 5′ end of the transposon, and the cellular splicing machinery can then create a usable poly(A)+ mRNA from the transposon alone, or by splicing to a cryptic downstream exon (see Figure 2). In the PBac enhancer traps we analyzed, transcription initiated in the first (noncoding) exon of a gene, and splicing produced GFP-coding mRNAs that terminated within the transposon. The enhancer trap lines therefore produce GFP that is not fused to endogenous proteins. This was confirmed by Western blot analysis of several enhancer traps; proteins detected with GFP antibodies were all ∼40 kDa in molecular weight. Another hallmark of this type of enhancer trap is accumulation of GFP in nuclei. For the P-element enhancer traps this is explained by the presence of a nuclear localization signal in the transposase peptide fused to GFP. The mechanism for nuclear targeting of the PBac-derived proteins is not known. While they are clearly not protein traps, the enhancer traps are still very useful since they provide accurate readouts of transcriptional activity.
Among the lines with insertions upstream of the coding sequence of a gene, we did find cases where the GFP-coding sequence was spliced to an annotated genomic coding exon and produced a fusion protein with GFP linked to the amino terminus. These have been reclassified as protein traps. However, the vast majority of the predicted enhancer trap lines had nuclear expression patterns and small GFP-containing proteins, suggesting that N-terminal fusion of GFP to endogenous proteins is uncommon in the lines we analyzed. Therefore, the majority of insertions that lie upstream of the coding region of an annotated gene likely represent enhancer traps.
This work underscores the importance of careful vector design. A number of constructs (protein traps, RNAi, and genomic rescuing fragments) contain exons that must be spliced for proper expression. Attempts should be made to avoid the possibility of splicing unintended portions of the transposon into the desired mRNAs. One easy way to do this may be to put genes in the reverse orientation of the transposable element used. It should be noted, however, that although the possibility of P-element exon 0 being spliced to the B frame of GFP could have been predicted, the splicing that occurred in the PBac 5′ end could not. This splicing pattern does not reflect any splicing that normally occurs in PBac.
We classified 125 lines, representing 82 genes, as enhancer opposite on the basis of the orientation of the insertions and proximity to an annotated gene. Although we did not analyze RNA from these lines, two pieces of evidence suggest they are also acting as enhancer traps. First, as with the enhancer traps, the enhancer opposites are overrepresented in the YB and YD lines, both of which we have shown include many lines that produce GFP-encoding mRNAs (see above). Second, 10 of 15 lines we examined on Western blots had GFP-positive bands ∼40 kDa. Therefore, expression of GFP in these lines is in response to enhancers that may or may not be associated with the gene they are inserted in.
Finally, we classified 128 lines as no gene because their insertions were located >500 nucleotides from any annotated gene. Although these could have been picking up unannotated genes, our evidence suggests they are acting as enhancer traps. The lines represent 75 different locations in the genome, 7 of which had at least two independent insertions. Of 19 lines analyzed by Western blotting, 17 had GFP-positive bands ∼40 kDa. One of the sites with multiple independent insertions is between CG6301 and mir-8 where we found 25 PBac insertions in the plus orientation among the YD lines and five independent P-element insertions in the minus orientation among the P, G, ZCL, and YB lines. RNA analysis of two of the P-element insertions showed that the lines were splicing into the same cryptic exon. Such hot spots could indicate that the genome contains strong transcriptional control sequences far removed from known transcription units (Manak et al. 2006). Alternatively, they may point to the positions of short noncoding RNA genes. However, very few of the no gene lines were lethal (14% compared to 42% lethality overall), suggesting that associated genes are not essential.
Mutagenicity and distribution of protein and enhancer traps:
Our screens produced higher mutagenicity rates than previously reported screens, as measured by homozygous lethality. Overall, 42% of the independent insertions in the YB, YC, and YD lines were associated with lethality (average of YB, YC, and YD lines; see Table 4). The P-element insertions (YB and YC) were 32% lethal and PBac insertions (YD) were 47% lethal. These results can be compared to earlier studies that reported lethality rates ranging from 11–13% using P elements (Bellen 1999) to 25–30% using SUPor-P transposons (Roseman et al. 1995). A PBac screen reported by Horn et al. (2003) resulted in 7.6% lethality. Finally, large collections of P-element (8300) and PBac element (18,000) insertions made recently were found to be associated with 17 and 22% lethality, respectively (Thibault et al. 2004).
We expected that protein trap insertion within introns would be less mutagenic than average since the insertion sites would not disrupt sequences around the transcription start site. We investigated this by comparing the rate of lethality of genes with enhancer traps (insertions upstream of the coding region) to that of genes with protein traps. Indeed, we did find that the genes we isolated with enhancer traps had higher lethality, 53%, than the genes with protein traps, 36% (Table 6). However, we found that about one-third of the lethal protein traps were viable over a deficiency, so the actual lethality of the lines is lower. Nonetheless, the high rate of lethality in the protein trap lines suggests that insertion of GFP into proteins or disruptions of normal splicing patterns can be deleterious. Still, the majority of the GFP-tagged fusion proteins retained specific localization patterns, showing that the proteins are likely folded properly and can localize appropriately.
P elements are known to have insertion hotspots, and the hotspots vary from screen to screen perhaps depending on the type of element and its starting location (Bellen et al. 2004). We noted evidence of different hotspots depending on what stage of development is screened for GFP+ insertions. We sequenced 25 independent insertions in the Dek gene among the G and ZCL lines made by Morin et al. (2001) (Table 5). However, we did not recover any insertions in Dek in our screens even though we used the same starting elements. This may be because Dek is highly expressed in first instar larvae, which were screened by Morin et al. (2001), and not in embryos, which we screened using the embryo sorter. Conversely, we isolated 11 insertions in extra macrochaetae (emc) by embryo screening, while no insertions in this gene were found by Morin et al. (2001). This indicates that it will be necessary to screen multiple stages of development to capture traps in all genes.
Lessons learned:
The automated embryo sorter was very effective in recovering rare events that led to the expression of GFP from the constructs we used. We found a wide range of expression levels during oogenesis among the lines we examined, including very faint, but specific expression patterns. This suggests that the sorter was able to detect embryos with a similarly wide range of expression levels, and we were able to capture events that produced low levels of fluorescence.
As discussed above, the cellular splicing machinery is capable of splicing the GFP exon to both cryptic and real exons in the genome and in the transposon ends. Therefore, care must be taken when designing traps that depend on splicing for expression. Known splice donor and acceptor sites in the transposons should be mutated so they are nonfunctional. In the configurations we tested, the white gene is the better marker to use. We did not find examples of white DNA incorporated into mRNAs, while there was a cryptic exon used from the yellow gene in the YD PBac screen.
The PBac-based trap construct we used was less effective than the P-element construct. The rates of producing GFP-positive insertions were comparable; however, expression of GFP was uniformly lower in PBac lines than in P-element lines. This may be because splicing of GFP into mRNAs of PBac protein traps is less efficient than that in P-element protein traps. The PBac construct also resulted in a higher frequency of lethal mutations, perhaps because of its relatively larger size compared to the P elements (8523 bp vs. 5961 bp).
Our work highlights the value of both multiple insertions in individual genes and insertions in genomic regions devoid of annotated genes. Many genes produce more than one protein isoform because of alternative splicing of exons or the use of more than one promoter. Determining the expression patterns of transcripts and protein isoforms from such a gene typically requires a combination of transcript-specific probes and isoform-specific antibodies. We described one example, insertions in the Imp gene, for which we found three distinct expression patterns reported by protein traps at different locations in the gene. Therefore, protein trapping holds great potential for deciphering isoform-specific expression patterns if sufficient fly stocks can be maintained. Similarly, insertions in DNA where no genes are annotated can be valuable in finding transcriptional control elements and characterizing their ability to activate transcription. Sufficiently dense coverage of the genome with protein traps and enhancer traps will contribute to complete annotation of the Drosophila transcriptome.
FlyTrap:
The greatest value of protein and enhancer trapping is in the information obtained from studying expression patterns. Having access to basic information about cell-specific expression and subcellular localization of tagged proteins produced by the gene provides an invaluable start toward understanding gene function. This is especially true for uncharacterized genes. Therefore, and perhaps most importantly, FlyTrap can serve as a central repository for image data collected from genes with traps. The initial image data set consists largely of ovarian tissue, with the addition of pupal wing images for some lines from Jeff Axelrod (Stanford University). However, FlyTrap can be used to collect expression data from other tissues so that a more complete view of gene expression patterns can be assembled.
Acknowledgments
We are very grateful to Xavier Morin and William Chia for sending us their protein trap lines and sharing unpublished information. We thank the following people for generous help with collecting virgins, sorting embryos, DNA sequencing, and collecting images: Stephanie Airoldi, Jonathan DiMaio, Michael Donnelly, Lynn Jones, Sangil Lee, Lorri Marek, Janine Mok, Brian O'Roak, Sharon Pozner-Moulis, Valeriy Rogulin, Sarah Woodfield, and Xinyan Yu. Remko De Knikker and Kei Cheung contributed to the development of the FlyTrap database. We thank Jeff Axelrod for pictures of pupal wings. We thank Martha Evans-Holm for determining flanking sequences by inverse PCR and Joseph W. Carlson and Robert Levis for alignment of flanking sequences to the genome sequence. Mapping experiments at Lawrence Berkeley National Laboratory and sequence curation done at the Carnegie Institution of Washington were supported by National Institutes of Health (NIH) grant GM067858 to Hugo J. Bellen. The COPAS Select embryo sorter was purchased with funds from an NIH shared instrument grant (RR16749) to L.C. S.M. was supported by a James Hudson Brown–Alexander Brown Coxe postdoctoral fellowship. This project was supported by NIH grant GM043301 to L.C.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas, D., A. E. Davidson, S. Sivasubbu, S. B. Hermanson, Z. Welle et al., 2004. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., 1999. Ten years of enhancer detection: lessons from the fly. Plant Cell 11: 2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec, Y., C. Marcaillou, X. Morin and A. Debec, 2003. Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil. Cytoskeleton 54: 217–225. [DOI] [PubMed] [Google Scholar]
- Bobinnec, Y., X. Morin and A. Debec, 2006. Shaggy/GSK-3beta kinase localizes to the centrosome and to specialized cytoskeletal structures in Drosophila. Cell Motil. Cytoskeleton 63: 313–320. [DOI] [PubMed] [Google Scholar]
- Bonin, C. P., and R. S. Mann, 2004. A piggyBac transposon gene trap for the analysis of gene expression and function in Drosophila. Genetics 167: 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak, M., S. Paterno, D. Lighthouse, J. Bachman, J. Planck et al., 2007. The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, J. P., S. M. Colicos and J. W. Belmont, 1999. Gene trap screening using negative selection: identification of two tandem, differentially expressed loci with potential hematopoietic function. Dev. Genet. 25: 49–63. [DOI] [PubMed] [Google Scholar]
- Cary, L. C., M. Goebel, B. G. Corsaro, H. G. Wang, E. Rosen et al., 1989. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia in transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172: 156–169. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805. [DOI] [PubMed] [Google Scholar]
- Clyne, P. J., J. S. Brotman, S. T. Sweeney and G. Davis, 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics 165: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, L., C. Berg, R. Kelley, D. McKearin and A. Spradling, 1989. Identifying and cloning Drosophila genes by single P element insertional mutagenesis. Prog. Nucleic Acid Res. Mol. Biol. 36: 99–109. [DOI] [PubMed] [Google Scholar]
- Cooley, L., E. Verheyen and K. Ayers, 1992. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 69: 173–184. [DOI] [PubMed] [Google Scholar]
- Dunn, A. K., and J. Handelsman, 1999. A vector for promoter trapping in Bacillus cereus. Gene 226: 297–305. [DOI] [PubMed] [Google Scholar]
- Gossler, A., A. L. Joyner, J. Rossant and W. C. Skarnes, 1989. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science 244: 463–465. [DOI] [PubMed] [Google Scholar]
- Grumbling, G., and V. Strelets, 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34: D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, D. D., D. Stein and L. M. Stevens, 2000. Investigating the function of follicular subpopulations during Drosophila oogenesis through hormone-dependent enhancer-targeted cell ablation. Development 127: 573–583. [DOI] [PubMed] [Google Scholar]
- Handler, A. M., and R. A. Harrell, 2nd, 1999. Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol. Biol. 8: 449–457. [DOI] [PubMed] [Google Scholar]
- Hirashima, M., A. Bernstein, W. L. Stanford and J. Rossant, 2004. Gene-trap expression screening to identify endothelial-specific genes. Blood 104: 711–718. [DOI] [PubMed] [Google Scholar]
- Horn, C., N. Offen, S. Nystedt, U. Hacker and E. A. Wimmer, 2003. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics 163: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jarvik, J. W., S. A. Adler, C. A. Telmer, V. Subramaniam and A. J. Lopez, 1996. CD-tagging: a new approach to gene and protein discovery and analysis. Biotechniques 20: 896–904. [DOI] [PubMed] [Google Scholar]
- Kelso, R. J., M. Buszczak, A. T. Quinones, C. Castiblanco, S. Mazzalupo et al., 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32: D418–D420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf, F., B. Rogina, Z. Jiang, P. S. Aronson and S. L. Helfand, 2002. Functional characterization and immunolocalization of the transporter encoded by the life-extending gene Indy. Proc. Natl. Acad. Sci. USA 99: 14315–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, T., S. Nagayoshi, A. Urasaki and K. Kawakami, 2006. Transposon-mediated gene trapping in zebrafish. Methods 39: 199–206. [DOI] [PubMed] [Google Scholar]
- Lee, J. K., E. Brandin, D. Branton and L. S. Goldstein, 1997. alpha-Spectrin is required for ovarian follicle monolayer integrity in Drosophila melanogaster. Development 124: 353–362. [DOI] [PubMed] [Google Scholar]
- Liao, G. C., E. J. Rehm and G. M. Rubin, 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97: 3347–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. G., Y. Chen and Q. Zhang, 2005. Amplification of genomic sequences flanking T-DNA insertions by thermal asymmetric interlaced polymerase chain reaction. Methods Mol. Biol. 286: 341–348. [DOI] [PubMed] [Google Scholar]
- Manak, J. R., S. Dike, V. Sementchenko, P. Kapranov, F. Biemar et al., 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 38: 1151–1158. [DOI] [PubMed] [Google Scholar]
- Morin, X., R. Daneman, M. Zavortink and W. Chia, 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare, K., and G. M. Rubin, 1983. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34: 25–35. [DOI] [PubMed] [Google Scholar]
- O'Kane, C. J., and W. J. Gehring, 1987. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 84: 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz et al., 1988. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics 118: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al., 1995. A P-element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Sineshchekova, O. O., T. Kawate, O. V. Vdovychenko and T. N. Sato, 2004. Protein-trap version 2.1: screening for expressed proteins in mammalian cells based on their localizations. BMC Cell Biol. 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, T., and E. Burke, 2003. High-throughput TAIL-PCR as a tool to identify DNA flanking insertions. Methods Mol. Biol. 236: 241–272. [DOI] [PubMed] [Google Scholar]
- Singleton, K., and R. I. Woodruff, 1994. a The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev. Biol. 161: 154–167. [DOI] [PubMed] [Google Scholar]
- Singleton, K., and R. I. Woodruff, 1994. b The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev. Biol. 161: 154–167. [DOI] [PubMed] [Google Scholar]
- Skarnes, W. C., B. A. Auerbach and A. L. Joyner, 1992. A gene trap approach in mouse embryonic stem cells: the lacZ reported is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes Dev. 6: 903–918. [DOI] [PubMed] [Google Scholar]
- Skarnes, W. C., J. E. Moss, S. M. Hurtley and R. S. Beddington, 1995. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92: 6592–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. J., 1997. Mini-exon epitope tagging for analysis of the protein-coding potential of genomic sequence. Biotechniques 23: 116–120. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. M. Stern, I. Kiss, J. Roote, T. Laverty et al., 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92: 10824–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Tsubota, S., M. Ashburner and P. Schedl, 1985. P-element-induced control mutations at the r gene of Drosophila melanogaster. Mol. Cell. Biol. 5: 2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vied, C., N. Halachmi, A. Salzberg and J. I. Horabin, 2003. Antizyme is a target of sex-lethal in the Drosophila germline and appears to act downstream of hedgehog to regulate sex-lethal and cyclin B. Dev. Biol. 253: 214–229. [DOI] [PubMed] [Google Scholar]
- Wakefield, S., and G. Tear, 2006. The Drosophila reticulon, Rtnl-1, has multiple differentially expressed isoforms that are associated with a sub-compartment of the endoplasmic reticulum. Cell Mol. Life Sci. 63: 2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, R. E., IV, R. S. Lamb and R. G. Fehon, 1998. A conserved functional domain of Drosophila coracle is required for localization at the septate junction and has membrane-organizing activity. J. Cell Biol. 140: 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, J. E., M. Buszczak and S. Sayles, 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9: 675–685. [DOI] [PubMed] [Google Scholar]
- Wilson, C., R. K. Pearson, H. J. Bellen, C. J. O'Kane, U. Grossniklaus et al., 1989. P-element-mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 3: 1301–1313. [DOI] [PubMed] [Google Scholar]