Abstract
Studies of transcriptional gene silencing in Drosophila melanogaster suggest that most of chromosome 4 resembles pericentric heterochromatin. However, some modifiers of position-effect variegation, including chromosome 4 dosage and loss of SU(VAR)3-9, have different effects on silencing in pericentric vs. distal arm chromosome 4 heterochromatin, distinguishing these two heterochromatin types.
IN Drosophila melanogaster, DNA packaged as heterochromatin is located around centromeres, at telomeres, and throughout the small fourth chromosome. The fourth is considered to be entirely heterochromatic by criteria including late replication and lack of recombination. However, the distal arm (1.2 Mb) forms a banded structure in polytene chromosomes and appears to have interspersed heterochromatic and euchromatic domains (Haynes et al. 2004; Sun et al. 2004; Riddle and Elgin 2006). Pericentric, telomeric, and chromosome 4 domains all have the capacity to induce position-effect variegation (PEV), the transcriptional silencing of a reporter gene placed near heterochromatin. Stable heterochromatin formation requires products encoded by suppressors of variegation [Su(var)s]. In pericentric regions, antipodal dosage dependence is seen for HP1, HP2, SU(VAR)3-7, and SU(VAR)3-9 (a histone H3K9 methyltransferase, HMT), suggesting that these are core structural proteins of heterochromatin (Locke et al. 1988; Eissenberg et al. 1990; Reuter et al. 1990; Cleard et al. 1997; Shaffer et al. 2006); HP1 interacts directly with the other three and binds H3 dimethylated at lysine 9 (H3K9me2) as well (reviewed by Schotta et al. 2003; Stephens et al. 2005). Telomere position effect (TPE) requires a distinct set of Mod(var)s and is generally unaffected by pericentric PEV modifiers (Wallrath and Elgin 1995; Cryderman et al. 1999; Boivin et al. 2003). The pericentric heterochromatin and distal fourth show colocalization of HP1, HP2, SU(VAR)3-7, and H3K9me2, suggesting a shared composition (Cleard et al. 1997; Shaffer et al. 2002; Haynes et al. 2004). Nonetheless, some differences have been observed.
While pericentric heterochromatin formation can spread hundreds of kilobases across rearrangement breakpoints (Zhimulev et al. 1988), organization of heterochromatin within the distal fourth chromosome appears to be more restricted with distinct interspersed heterochromatic and euchromatic domains (Sun et al. 2000). Heterochromatic silencing within the 200 kb surrounding variegating hsp70-white reporter 39C-12 appears to spread only ∼10 kb (Sun et al. 2004). In larvae homozygous for the Su(var)3-906 null allele, H3K9me2 is dramatically reduced in pericentric regions, accompanied by loss of HP1, but the distal fourth arm appears unaffected (Schotta et al. 2002), suggesting activity of a different HMT. To explore differences between pericentric and distal fourth chromosome heterochromatin, we have examined effects of fourth chromosome dosage and of specific Mod(var)s on silencing (PEV) in the two domains.
Chromosome 4 dosage specifically modifies PEV of chromosome 4 reporters:
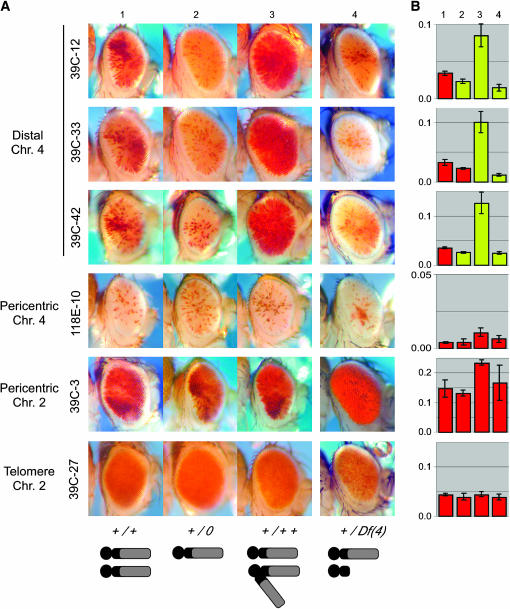
The amount of heterochromatin within the nucleus has an impact on PEV (Ashburner et al. 2004); additional heterochromatin is thought to increase competition for a fixed pool of heterochromatin components, destabilizing heterochromatic silencing at a variegating gene. Extra heterochromatic material introduced by a Y or attached X chromosome suppresses PEV of pericentric and chromosome 4 hsp70-white reporters (Wallrath and Elgin 1995), indicating shared heterochromatin components. Here we use an attached fourth chromosome [C(4)RM] to manipulate dosage of the largely heterochromatic chromosome 4, analyzing PEV in haplo-, diplo-, and triplo-4 flies. Silencing of distal chromosome 4 reporters (39C-12, 39C-33, and 39C-42) is consistently enhanced in a haplo-4 background and greatly suppressed in a triplo-4 background (Figure 1). In contrast, haplo-4 and triplo-4 genotypes have no statistically significant impact for characteristic pericentric reporters (118E-10 and 39C-3). To distinguish the dosage effect of the distal arm from that of pericentric fourth heterochromatin, we used terminal deletion D(4)B2-7AT, which eliminates most of the distal fourth, leaving the centromere region proximal to RpS3A intact (Sousa-Neves et al. 2005). The impact of this deletion on PEV is similar to that seen in haplo-4 flies (Figure 1); thus, the chromosome 4-specific haplo-enhancer effect can be attributed to the banded distal portion of the fourth. The dosage effect of chromosome 4 suggests competition for a fixed pool of heterochromatin components specific to that domain. However, we cannot rule out the possibility of an effect from changes in a chromosome 4-specific modifier of PEV either on the fourth itself or elsewhere in the genome.
Figure 1.—
Increased copies of chromosome 4 suppress PEV of chromosome 4 distal arm P-insert reporters. Variegating P[hsp26-plant, hsp70-white] reporters within the distal portion of chromosome 4 (39C-12, 39C-33, and 39C-42 at polytene divisions 102B, 102D, and 102F, respectively), pericentric heterochromatin (118E-10 and 39C-3, bases of chromosomes 4 and 2L, respectively), or telomeric heterochromatin (39C-27, chromosome 2) (described in Wallrath and Elgin 1995; Sun et al. 2000, 2004) were introduced into backgrounds with varying chromosome 4 dosage. Males were photographed 3–5 days postelcosion. (A) Columns (1–4) from left to right: 1, diplo-4 (+/+); 2, haplo-4 (+/0); 3, triplo-4 [C(4)eyR/+]; and 4, fourth chromosome deficiency [+/Df(4)B2-7AT]. Genotypes and graphic depictions of chromosome 4 karyotypes are shown below each column of photographs. (B) Quantitative measurements of eye pigment from the progeny shown in the photographs (same order from left to right) were carried out as described (Sun et al. 2004). Bars indicate means of three to five samples (five adult males each) with the standard error shown as a vertical goalpost. Mean values that are significantly different from diplo-4 (P < 0.05) are highlighted in yellow. Enhancement of PEV in flies carrying distal fourth chromosome reporters in the presence of a distal chromosome 4 deletion demonstrates that the haplo-enhancer effect can be attributed to the banded distal portion of chromosome 4. Drosophila cultures were raised on cornmeal sucrose-based medium at 25° as described (Shaffer et al. 1994). Stock BL1785 [C(4)RM, ci1 eyR/0] (Bloomington Drosophila Stock Center) is the source of the attached fourth chromosome for the line y w67c23; C(4)RM, ci1 eyR/0 used in our analysis. Stock y; Df(4)B2-7AT/ciD spapol was provided by R. Sousa-Neves (described in Sousa-Neves et al. 2005). In each experiment, PEV reporter lines were crossed to host stock y w67c23 as a wild-type control. To produce haplo- and triplo-4 flies, virgin females carrying the PEV reporter were crossed with males carrying an attached fourth chromosome [y w67c23; C(4)RM, ci1 eyR/0]. The fourth chromosome homologs segregate together, resulting in haplo-4 “minute” +/0 and triplo-4 C(4)RM, ci1 eyR/+ siblings.
Variegating reporters in distal chromosome 4 and pericentric regions respond differently to certain modifiers of PEV:
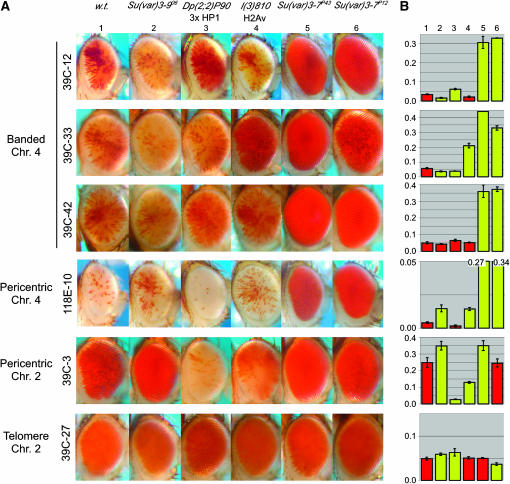
Mod(var) mutations were used to investigate which components distinguish pericentric and distal fourth PEV. Recent studies suggest separable effects of SU(VAR)3-7 subdomains on PEV within different heterochromatic regions (Bushey and Locke 2004; Jaquet et al. 2006). We find that mutations in both the C-terminal and Zn finger domains [Su(var)3-7P43 and Su(var)3-7P12, respectively] greatly diminish silencing of pericentric and distal fourth chromosome P reporters (Figure 2). Thus, SU(VAR)3-7 appears to be a common factor required for heterochromatin formation in both domains. Loss of histone variant H2Av [l(3)810] (van Daal and Elgin 1992) is reported to suppress PEV in In(1)wm4, suggesting a critical role for H2Av in heterochromatin (Swaminathan et al. 2005). The effect of l(3)810 varies at each insert we have tested (suppression, enhancement, or no significant alteration of PEV), suggesting that H2Av may contribute to locus-specific differences within pericentric and distal fourth domains (Figure 2). While all reporters used here show loss of silencing with a loss of HP1 (Wallrath and Elgin 1995; Sun et al. 2000; E. Gracheva, unpublished observations), increased HP1 dosage [Dp(2;2)P90] shows locus-specific effects on distal fourth chromosome PEV. Surprisingly, PEV at insert 39C-12 is suppressed by an increase of HP1; previous studies have shown that variegation at 39C-12 is also suppressed by a loss of HP1 (Sun et al. 2000). Increased quantities of HP1 might selectively reinforce robust heterochromatin formation at certain loci and effectively deprive other loci such as 39C-12 of heterochromatin-forming components. Su(var)3-906 is the only mutation observed that appears to distinguish pericentric and distal chromosome 4 heterochromatin domains consistently. Loss of SU(VAR)3-9 suppresses PEV at the pericentric reporters but fails to suppress PEV at the distal chromosome 4 reporters. Interestingly, PEV of two distal fourth reporters is enhanced by the loss of SU(VAR)3-9. Since HP1 localization at pericentric heterochromatin is dependent upon SU(VAR)3-9 (Schotta et al. 2002), loss of this protein destabilizes pericentric heterochromatin, likely liberating heterochromatic components, including HP1 and SU(VAR)3-7, possibly for assembly at certain chromosome 4 loci, enhancing silencing there. Spreading of heterochromatin in pericentric regions may rely upon the simultaneous recognition of H3K9me2 and recruitment of SU(VAR)3-9 methyltransferase by HP1 (reviewed by Elgin and Richards 2002). Since distal fourth chromosome PEV, H3K9 dimethylation, and HP1 localization do not require SU(VAR)3-9, heterochromatin formation in this domain may require a different HMT and therefore utilize a profoundly different heterochromatin-spreading mechanism. The unidentified HMT might play a role in the observed chromosome 4-specific dosage effect. The distal arm of chromosome 4 thus represents a distinct compartment of heterochromatin.
Figure 2.—
A comparative genetic analysis of fourth chromosome vs. pericentric PEV. (A) Photographs of male progeny from crosses between females carrying a PEV reporter (as in Figure 1) and the following males: 1, y w67c23 (wild type); 2, Su(var)3-906/Su(var)3-906 [SU(VAR)3-9 null (Reuter et al. 1986)]; 3, Dp(2;2)P90/CyO [two doses of HP1 (Wustmann et al. 1989), provided by J. C. Eissenberg]; 4, l(3)810/TM6 Tb [haplo-deficient H2Av (van Daal and Elgin 1992)]; 5, Su(var)3-7P43/TM6 [G416E point mutation in Zn finger 4 of SU(VAR)3-7 (Bushey and Locke 2004)]; 6, Su(var)3-7P12/TM6 [C-terminal deletion of SU(VAR)3-7 (Bushey and Locke 2004)]. (B) Eye pigment measurements and photographs of progeny without the balancer chromosome were carried out as described in the Figure 1 legend. Mean values that are significantly different from those of wild type (P < 0.05) are highlighted in yellow. Responses to a loss of SU(VAR)3-9 distinguish the fourth chromosome distal arm from the pericentric heterochromatin.
Acknowledgments
This work was funded by National Institutes of Health grant GM068388 to S.C.R.E.
References
- Ashburner, M., 2004. Position effect variegation, pp. 1007–1038 in Drosophila: A Laboratory Handbook, edited by M. Ashburner, K. Golic and S. W. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Boivin, A., C. Gally, S. Netter, D. Anxolabehere and S. Ronsseray, 2003. Telomeric associated sequences of Drosophila recruit polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164: 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey, D., and J. Locke, 2004. Mutations in Su(var)205 and Su(var)3–7 suppress P-element-dependent silencing in Drosophila melanogaster. Genetics 168: 1395–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleard, F., M. Delattre and P. Spierer, 1997. SU(VAR)3–7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J. 16: 5280–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman, D. E., E. J. Morris, H. Biessmann, S. C. Elgin and L. L. Wallrath, 1999. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 18: 3724–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., T. C. James, D. M. Foster-Hartnett, T. Hartnett, V. Ngan et al., 1990. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 87: 9923–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin, S. C. R., and E. Richards, 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500. [DOI] [PubMed] [Google Scholar]
- Haynes, K. A., B. A. Leibovitch, S. H. Rangwala, C. Craig and S. C. Elgin, 2004. Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harbor Symp. Quant. Biol. 69: 267–272. [DOI] [PubMed] [Google Scholar]
- Jaquet, Y., M. Delattre, J. Montoya-Burgos, A. Spierer and P. Spierer, 2006. Conserved domains control heterochromatin localization and silencing properties of SU(VAR)3–7. Chromosoma 115: 139–150. [DOI] [PubMed] [Google Scholar]
- Locke, J., M. A. Kotarski and K. D. Tartof, 1988. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics 120: 181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, G., R. Dorn, G. Wustmann, B. Friede and G. Rauh, 1986. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol. Gen. Genet. 202: 481–487. [Google Scholar]
- Reuter, G., M. Giarre, J. Farah, J. Gausz, A. Spierer et al., 1990. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature 344: 219–223. [DOI] [PubMed] [Google Scholar]
- Riddle, N. C., and S. C. R. Elgin, 2006. The dot chromosome of Drosophila: insights into chromatin states and their change over evolutionary time. Chromosome Res. 14: 405–416. [DOI] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann et al., 2002. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta, G., A. Ebert, R. Dorn and G. Reuter, 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell. Dev. Biol. 14: 67–75. [DOI] [PubMed] [Google Scholar]
- Shaffer, C. D., J. M. Wuller and S. C. Elgin, 1994. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 44: 99–108. [DOI] [PubMed] [Google Scholar]
- Shaffer, C. D., G. E. Stephens, B. A. Thompson, L. Funches, J. A. Bernat et al., 2002. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc. Natl. Acad. Sci. USA 99: 14332–14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, C., G. Cenci, B. Thompson, G. E. Stephens, E. Slawson et al., 2006. The large isoform of Drosophila melanogaster heterochromatin protein 2 plays a critical role in gene silencing and chromosome structure. Genetics 174: 1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Neves, R., T. Lukacsovich, C. M. Mizutani, J. Locke, L. Podemski et al., 2005. High-resolution mapping of the Drosophila fourth chromosome using site-directed terminal deficiencies. Genetics 170: 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, G. E., E. E. Slawson, C. Craig and S. C. R. Elgin, 2005. Interaction of heterochromatin protein 2 with HP1 defines a novel HP1 binding domain. Biochemistry 44: 13394–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F. L., M. H. Cuaycong, C. A. Craig, L. L. Wallrath, J. Locke et al., 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. USA 97: 5340–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F. L., K. Haynes, C. L. Simpson, S. D. Lee, L. Collins et al., 2004. cis-acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24: 8210–8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, J., E. M. Baxter and V. G. Corces, 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal, A., and S. C. Elgin, 1992. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 3: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath, L. L., and S. C. Elgin, 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263–1277. [DOI] [PubMed] [Google Scholar]
- Wustmann, G., J. Szidonya, H. Taubert and G. Reuter, 1989. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol. Gen. Genet. 217: 520–527. [DOI] [PubMed] [Google Scholar]
- Zhimulev, I. F., E. S. Belyaeva, A. V. Bgatov, E. M. Baricheva and I. E. Vlassova, 1988. Cytogenetic and molecular aspects of position effect variegation in Drosophila melanogaster. II. Peculiarities of morphology and genetic activity of the 2B region in the T(1;2)dorvar7 chromosome in males. Chromosoma 96: 255–261. [DOI] [PubMed] [Google Scholar]