Abstract
DNA replication initiation in S. cerevisiae is promoted by B-type cyclin-dependent kinase (Cdk) activity. In addition, once-per-cell-cycle replication is enforced by cyclin-Cdk-dependent phosphorylation of the prereplicative complex (pre-RC) components Mcm2-7, Cdc6, and Orc1-6. Several of these controls must be simultaneously blocked by mutation to obtain rereplication. We looked for but did not obtain strong evidence for cyclin specificity in the use of different mechanisms to control rereplication: both the S-phase cyclin Clb5 and the mitotic cyclins Clb1–4 were inferred to be capable of imposing ORC-based and MCM-based controls. We found evidence that the S-phase cyclin Clb6 could promote initiation of replication without blocking reinitiation, and this activity was highly toxic when the ability of other cyclins to block reinitiation was prevented by mutation. The failure of Clb6 to regulate reinitiation was due to rapid Clb6 proteolysis, since this toxic activity of Clb6 was lost when Clb6 was stabilized by mutation. Clb6-dependent toxicity is also relieved when early accumulation of mitotic cyclins is allowed to impose rereplication controls. Cell-cycle timing of rereplication control is crucial: sufficient rereplication block activity must be available as soon as firing begins. DNA rereplication induces DNA damage, and when rereplication controls are compromised, the DNA damage checkpoint factors Mre11 and Rad17 provide additional mechanisms that maintain viability and also prevent further rereplication, and this probably contributes to genome stability.
DNA replication must take place only once per cell cycle in eukaryotes. This mechanism is controlled at the level of prereplicative complex (pre-RC) formation and origin firing. The pre-RC is assembled at replication origins. The origin recognition complex (Orc1-6) is bound to DNA replication origins throughout the cell cycle in Saccharomyces cerevisiae (Diffley et al. 1994). The Cdc6, Cdt1, and the mini-chromosome maintenance proteins (MCM) Mcm2–7 are sequentially recruited to the origin to form the pre-RC (Cocker et al. 1996; Aparicio et al. 1997; Devault et al. 2002; Tanaka and Diffley 2002; Tanaka et al. 1997). The assembled pre-RC further recruits Cdc45, DNA polymerase α, Sld2, and other factors required for initiation and elongation (Bell and Dutta 2002). Activation of two kinases, Dbf4-Cdc7 and cyclin-dependent kinases (CDK), coupled with B-type cyclins are necessary to form replisomes at two nascent replication forks and to initiate DNA replication (Bell and Dutta 2002).
Multiple mechanisms prevent cells from starting a second round of initiation by inhibiting pre-RC formation (Broek et al. 1991; Hayles et al. 1994; Dahmann et al. 1995). In S. cerevisiae, phosphorylation of MCMs by Cln-Cdk1 and Clb-Cdk1 kinases causes their exclusion from the nucleus (Labib et al. 1999; Nguyen et al. 2000; Liku et al. 2005). Phosphorylation of Orc2 and Orc6 by Clb-Cdk1 is thought to prevent efficient binding of other pre-RC subunits (Nguyen et al. 2001). Binding of Clb5 to Orc6 via its RXL cyclin-binding motif contributes to prevention of rereplication (Wilmes et al. 2004). Phosphorylation targets of Clb5-Cdk1 likely include Orc6 itself as well as Orc2 and Orc1 (Nguyen et al. 2001; Archambault et al. 2004; Wilmes et al. 2004). Finally, Cdc6 is inhibited by multiple mechanisms through its N terminus. N-terminal phosphorylation of Cdc6 promotes its degradation by the proteasome (Drury et al. 1997; Elsasser et al. 1999; Calzada et al. 2000; Drury et al. 2000), and binding of Cdc6p to mitotic cyclins inactivates Cdc6 for origin loading; Cdc6-cyclin binding is dependent on N-terminal phosphorylation of Cdc6 (Mimura et al. 2004). Additionally, Cdc6 has an N-terminal nuclear localization signal, which is critical for its degradation (Luo et al. 2003). When multiple disruptions of these mechanisms are combined, cells undergo extensive rereplication (Nguyen et al. 2001; Wilmes et al. 2004). In other eukaryotes, similar but distinct mechanisms combine to control rereplication (Kearsey and Cotterill 2003).
We recently reported that a synthetic genetic array (SGA) analysis identified genes required for viability when rereplication controls were partially compromised by combining the ORC6-rxl mutation blocking Clb5 binding to Orc6, and the CDC6ΔNT mutation, an N-terminal truncation resulting in abrogation of Cdk control of Cdc6 activity (see above). This background is initially slow growing but viable; we searched the haploid viable deletion set for deletions that would result in tight inviability in an ORC6-rxl CDC6ΔNT background. We found two major clusters of genes: one related to DNA damage response and one related to cell-cycle regulation (Archambault et al. 2005b). Further experiments suggested that the DNA damage response cluster was detected because cells partially compromised for rereplication control suffered double-strand breaks and required DNA damage surveillance mechanisms for viability (Archambault et al. 2005b). Rereplication can induce extensive DNA damage and could lead to genome instability (Archambault et al. 2005b; Green and Li 2005).
Here, we investigate the significance of the cell-cycle regulation cluster detected in our SGA analysis.
MATERIALS AND METHODS
Yeast strain construction:
Strain list was provided in Table 1. Standard methods were used for mating, tetrad analysis, and transformations. The ORC6-rxl allele has mutations at R178A and L180A; these mutations strongly reduce the specific interaction between Clb5 and the ORC complex (Wilmes et al. 2004). The ORC6-ps allele has phosphorylation site mutations at S106A, S116A, S123A, and T146A (Wilmes et al. 2004). GAL-CDC6ΔNT-HAs (Δ2-48) (single copy) and GAL-CDC6ΔNT-HAm (multiple copy) were constructed by transformation using linearized RS305-based GAL-CDC6Δ2-48-HA plasmid (Wilmes et al. 2004). These constructs allow strong unregulated expression and accumulation of Cdc6. In all strains with GAL-CDC6 constructs, the wild-type CDC6 gene was also present to allow viability on glucose medium where the GAL-CDC6 constructs were not expressed. The MCM7-NLS allele, allowing cell-cycle-constitutive nuclear residence of the Mcm complex, was described in Nguyen et al. (2001). CLB6Δ3P has three mutations at S6A, T39A, and S147A, stabilizing the protein by blocking Cdk phosphorylation and Skp1/Cull/F-Box protein (SCF)-dependent degradation (Jackson et al. 2006).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| RUY121 | Wild type; MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 |
| RUY139 | MATα sic1∷HIS3 |
| RUY275 | MATα orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm |
| RUY384 | MAT? orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm sic1∷HIS3 |
| RUY385 | MAT? orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm sic1∷HIS3 clb5∷URA3 clb6∷KanMX |
| RUY386 | MATaorc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm sic1∷HIS3 clb6∷KanMX |
| RUY387 | MATα orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm clb5∷URA3 clb6∷KanMX |
| RUY140 | MATα cdh1∷LEU2 |
| RUY263 | MATα URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY227 | MATα clb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY174 | MATaclb5∷URA3 clb6∷KanMX URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY228 | MATaMCM7-NLS URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY179 | MATaMCM7-NLS clb5∷URA3 clb6∷KanMX URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY170 | MATα orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY181 | MATα orc6∷HIS3∷LEU2∷ORC6-ps,rxl clb5∷URA3URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY184 | MATaorc6∷HIS3∷LEU2∷ORC6-ps,rxl clb5∷URA3 clb6∷KanMX URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY169 | MATα MCM7-NLS orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY191 | MATaMCM7-NLS orc6∷HIS3∷LEU2∷ORC6-ps,rxl clb5∷URA3 clb6∷KanMX URA3∷GAL-CDC6ΔNT-HAm DDC2-GFP∷TRP1 |
| RUY325 | MATα MCM7-NLS clb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm |
| RUY326 | MATα clb5∷URA3 URA3∷ GAL-CDC6ΔNT-HAm MCM7-NLS cdh1∷LEU2 |
| RUY172 | MATaclb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm |
| RUY329 | MATaclb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm cdh1∷LEU2 |
| RUY332 | MATaclb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm orc6∷HIS3∷LEU2∷ORC6-ps,rxl |
| RUY334 | MATaclb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm orc6∷HIS3∷LEU2∷ORC6-ps,rxl cdh1∷LEU2 |
| RUY415 | MATaMCM7-NLS clb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm CLB6Δ3p-HA∷URA3 |
| RUY435 | MATarad17∷KanMX clb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm |
| RUY436 | MATarad17∷KanMX orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm |
| RUY437 | MATα rad17∷KanMX clb5∷URA3 orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm |
| RUY438 | MAT? mre11∷KanMX clb5∷URA3 URA3∷GAL-CDC6ΔNT-HAm |
| RUY439 | MAT? mre11∷KanMX orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm |
| RUY440 | MAT? mre11∷KanMX clb5∷URA3 orc6∷HIS3∷LEU2∷ORC6-ps,rxl URA3∷GAL-CDC6ΔNT-HAm |
| RUY299 | MATα orc6∷HIS3∷LEU2∷ORC6-ps,rxl MCM7-NLS URA3∷GAL-CDC6ΔNT-HAm rad17∷KanMX |
| RUY302 | MAT? orc6∷HIS3∷LEU2∷ORC6-ps,rxl MCM7-NLS URA3∷GAL-CDC6ΔNT-HAm mre11∷KanMX |
| RUY303 | MATaorc6∷HIS3∷LEU2∷ORC6-ps,rxl MCM7-NLS URA3∷GAL-CDC6ΔNT-HAm mre11∷KanMX rad17∷KanMX |
| RUY280 | MATaclb1 clb2(ts) clb3∷TRP1 clb4∷his3∷KanMX URA3∷GAL-CDC6ΔNT-HAm orc6∷HIS3∷LEU2∷ORC6-ps,rxl |
| Alleles used in this study | Description and references |
| ORC6-rxl | Linealized pRS405 (LEU marked)-based ORC6-rxl plasmid was integrated into orc6∷HIS3MX strain. ORC6-rxl contains mutations in RXL motif, which interfere with Clb5 binding (Wilmes et al. 2004). |
| GAL-CDC6ΔNT-HAs | Deletion of N-terminal region of CDC6 allows its stable expression due to insufficient phosphorylation by CDKs. The plasmid was kindly provided by J. Diffley. The single integration of GAL-CDC6Δ2-48-HA was confirmed by Southern blotting. Endogenous copy of CDC6 exists in addition to GAL-CDC6Δ2-48 in the strain (Archambault et al. 2005a,b). |
| ORC6-ps,rxl | ORC6-ps,rxl contains ORC6 Cdk phosphorylation site mutations in addition to ORC6-rxl. pRS405-based ORC6-ps,rxl plasmid was linealized and integrated into the orc6∷HIS3MX strain (Wilmes et al. 2004). |
| GAL-CDC6ΔNT-HAm | Multiple copy of GAL-CDC6Δ2-48-HA was integrated on the basis of Southern blotting. Otherwise the feature of this strain is as the same as that of GAL-CDC6ΔNT-HAs (Wilmes et al. 2004). |
| MCM7-NLS | Endogenous copy of Mcm7 was fused at its C terminus to two tandem copies of the SV40 nuclear localization signal allowing constitutive nuclear localization of Mcm7 (Nguyen et al. 2001). |
| CLB6Δ3P | Three serines or threonines of the phosphorylation sites in Clb6 were mutated to alanine, and the resulting CLB6Δ3P was expressed from the endogenous CLB6 promoter (Jackson et al. 2006). The CLB6Δ3P stabilizes Clb6p and escapes from SCF-dependent degradation. |
Serial dilution:
Each strain was grown to stationary phase in 3 ml YPD. After the cell concentration was normalized on the basis of OD measurement, 5 μl of 10-fold serial dilutions were spotted onto YEP-D or YEP-G (glucose or galactose) plates. The plates were incubated for 2 days at 30°.
Rereplication assay:
For induction of rereplication, cells were grown in liquid YEP-D (glucose) media overnight, washed, and transferred to YEP-R (raffinose) for 8 hr. GAL-CDC6ΔNT-HAm was then induced by adding 3% galactose (final concentration) for 4 hr.
Microscopy and DNA flow cytometry analysis:
Ddc2-GFP foci were observed under a DeltaVision microscope as described (Archambault et al. 2005b). DNA flow cytometry analysis was performed as described (Epstein and Cross 1992).
RESULTS
Genetic interactions between rereplication controls and the cell-cycle regulators SIC1 and CLB5:
The RXL motif of Orc6 interacts with Clb5 to locally prevent origin refiring (Wilmes et al. 2004). Combining a mutation disrupting the Orc6-Clb5 interaction (ORC6-rxl) with another mutation stabilizing Cdc6 and preventing its sequestration by Clb2-Cdk1 (CDC6ΔNT) causes reduced viability and proliferation. Similarly, deletion of CLB5 in a CDC6ΔNT background is lethal in tetrad analysis (Wilmes et al. 2004), and clb5 GAL-CDC6ΔNT cells were nearly inviable on galactose plates (YEP-G) (Figure 1, A and B). In both experiments, removing Clb5 had a more profound effect than removing the Orc6-RXL (Clb5-binding) in the CDC6ΔNT background. Clb5 could engage in residual binding with Orc6-rxl or could have other functions in preventing rereplication that are independent of Orc6 binding (discussed below).
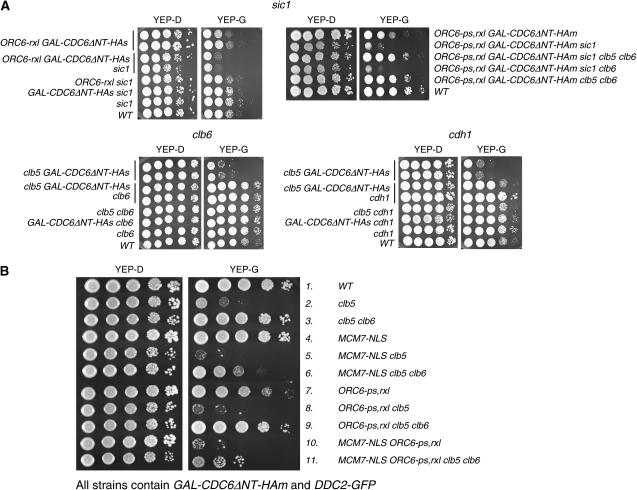
Figure 1.—
The survival of cells with disrupted rereplication controls depends on cell-cycle regulators. (A) ORC6-rxl GAL-CDC6ΔNT-HAs cells (top left) rely on SIC1 for their limited survival when GAL-CDC6ΔNT-HAs is induced by galactose. Ten-fold serial dilutions on galactose-containing or glucose-containing plates (YEP-G or YEP-D) were performed. Deletion of SIC1 in ORC6-ps,rxl GAL-CDC6ΔNT-HAm caused synthetic lethality. The lethality was rescued by deleting both CLB5 and CLB6 (top right). The viability of clb5 GAL-CDC6ΔNT-HAs cells (bottom) is rescued by deletion of CLB6 or CDH1 on YEP-G. (B) Viability of various strains with disruptions of mechanisms preventing rereplication in the presence or absence of S-phase cyclins. All strains contain GAL-CDC6ΔNT-HAm and DDC2-GFP (analyzed in Figure 2). Cell viability was tested as in A.
In our previous study, we conducted a genomewide screen using the ORC6-rxl GAL-CDC6ΔNT-HAs strains. (GAL-CDC6ΔNT-HAs is a single copy of GAL-CDC6; in some experiments, we also use a multicopy GAL-CDC6ΔNT-HAm allele, as described by Archambault et al. 2005b.) We identified a cluster of genes related to DNA damage checkpoints that are strongly required for viability in this background (Archambault et al. 2005b). In addition, we detected a cluster of genes that were related to cell-cycle regulation, including CLB5 and SIC1 (Figure 1A).
Deletion of SIC1 in the ORC6-rxl GAL-CDC6ΔNT-HAs background markedly decreased cell viability on YEP-G (Figure 1A, top left). The main function of Sic1 is to inhibit Clb5/Clb6-Cdk in late G1 (Mendenhall 1993; Schwob et al. 1994). Since the Clb5-Orc6 interaction is already disabled in the ORC6-rxl GAL-CDC6ΔNT-HAs background, lack of Sic1 in this context may be toxic through a mechanism that is independent of this interaction. We speculated that this is due to an elevated or a premature activation of Clb5,6 activity. The sic1 deletion in ORC6-ps,rxl GAL-CDC6ΔNT-HAm [ORC6-ps,rxl lacks both the RXL sequence and the Cdk phosphorylation sites described in Nguyen et al. (2001) and Wilmes et al. (2004)] also reduced viability (Figure 1A, top right). Consistent with the idea that premature Clb5,6 activity was responsible for lethality of ORC6-ps,rxl GAL-CDC6ΔNT-HAm sic1, the lethality was rescued by additional deletion of both CLB5 and CLB6 (Figure 1A, top right). We speculate that the possible mechanism of premature activation of Clb5,6 activity in the sic1 ORC6-ps,rxl GAL-CDC6ΔNT-HAm leads to initiation without blocking origin reloading (due to ORC6-ps,rxl mutation), leading to rereplication.
Induction of rereplication requires stabilized Cdc6 (CDC6ΔNT) in most genetic combinations tested in this study. This implies that degradation of Cdc6 plays a significant role in the inhibition of DNA rereplication. Interestingly, in tetrad analysis, deletion of CLB5 in the ORC6-ps,rxl sic1 background was lethal (even with wild-type CDC6 as the sole source of CDC6) (data not shown). Clb6p is responsible for this lethality, since additional deletion of CLB6 in the ORC6-ps,rxl sic1 clb5 background restored viability (Figure 1A, top right, YEP-D, third row).
Deletion of CLB6 or CDH1 rescues defects caused by clb5 CDC6ΔNT:
The lethality of the clb5 GAL-CDC6ΔNT-HAs strain in galactose was efficiently rescued by additional deletion of CLB6 (Figure 1A, bottom left). The genetic combinations that caused reduced viability involving perturbed rereplication control and CLB5 deletion (GAL-CDC6ΔNT clb5 sic1 and CDC6ΔNT clb5) were rescued by additional deletion of CLB6 (data not shown). All of these backgrounds share deregulation of Cdc6 and prevention of Clb5-Orc6 interaction by clb5 deletion. These results support the idea that in the presence of stabilized Cdc6, Clb6 is toxic when Clb5-based rereplication inhibition mechanisms are abolished. This implies that Clb5 has greater ability to block rereplication than Clb6, despite sequence similarity and the close evolutionary relationship of Clb5 and Clb6 (Schwob and Nasmyth 1993).
Cdh1 is known to target residual mitotic B-type cyclins (Clb1-4) for degradation early in the cell cycle (Visintin et al. 1997; Schwab et al. 2001; Wasch and Cross 2002). Deleting CDH1 in clb5 GAL-CDC6ΔNTs cells completely rescued their proliferation on YEP-G (Figure 1A, bottom right). Deletion of CDH1 in this background also rescued the rereplication phenotype determined by DNA flow cytometry analysis (see below). Consistently, inviability of clb5 CDC6ΔNT (endogenous gene replacement) or slow growth of ORC6-rxl CDC6ΔNT segregants in tetrad analysis was efficiently rescued by deletion of CDH1 (data not shown). In the absence of Cdh1, mitotic cyclins are present at a nearly constitutive level through the cell cycle, including in early G1, most likely due in part to inheritance of cyclin from the previous cell cycle; levels later in the cell cycle are quite similar to levels in wild-type cells (Cross et al. 2002; Wasch and Cross 2002). Therefore, we attribute the effects of CDH1 deletion on replication control as being due to early presence of mitotic cyclins. Increasing the level of mitotic cyclins early in the cell cycle may improve the viability of clb5 CDC6ΔNT or ORC6-rxl CDC6ΔNT cells by eliminating a temporal gap between S-Clb-Cdk and M-Clb-Cdk activation. This could allow a stronger reinitiation block at the time of replication initiation, compensating for the disruption of the Clb5-Orc6 interaction.
The absence of Cdh1 is likely to result in stabilization of several proteins in addition to B-type cyclins. To test whether an enhanced presence of mitotic cyclin in early S-phase could rescue clb5 CDC6ΔNT inviability, we placed CLB2 instead of CLB5 under the control of the CLB5 promoter (CLB5pCLB2, where CLB5 is absent) (Cross et al. 1999). CLB5pCLB2 CDC6ΔNT spores formed small colonies, while clb5 CDC6ΔNT spores were completely inviable (supplemental Table S1 at http://www.genetics.org/supplemental/). Thus two independent means of inducing early accumulation of mitotic cyclins relieved inviability of clb5 CDC6ΔNT.
As noted in our previous work (Archambault et al. 2005b), a concern with these experiments is that the mutations that we are using to deregulate rereplication control are in proteins (S-phase cyclins, pre-RC components) that also positively control replication, and some of the toxicity that we detect could be due to poor initial origin usage, rather than to rereplication. We tested this by assessing bulk DNA replication in synchronized cells and found at most modest defects in replication initiation (supplemental Figure S1 at http://www.genetics.org/supplemental/). Recently, Green et al. (2006) showed little or no replication initiation defects associated with ORC, MCM, and CDC6 mutations deregulating replication control by a microarray-based assay. Thus, overall we consider it unlikely that replication initiation defects contribute significantly to the results presented here.
Rereplication and accumulation of Ddc2-GFP DNA damage foci:
The previous results suggest that the relative timing of onset of S-phase cyclin and mitotic cyclin activities is crucial for viability in strains in which specific mechanisms preventing rereplication have been disrupted. To confirm this idea, we examined DNA rereplication directly by DNA flow cytometry and indirectly by accumulation of Ddc2-GFP (DNA damage foci) in the presence or absence of S-phase cyclin, accompanied by inactivation of various rereplication controls. We and others showed previously that Ddc2-GFP foci accumulate under rereplication conditions, even in the absence of overt rereplication detectable by DNA flow cytometry (Archambault et al. 2005b; Green and Li 2005). In the following sections, we describe how we used these assays to examine rereplication and accumulation of DNA damage in various backgrounds.
Clb5 blocks Clb6-dependent rereplication in cells expressing stable Cdc6:
DNA flow cytometry analysis showed an accumulation of cells with a signal increased to slightly more than 2C DNA content in clb5 GAL-CDC6ΔNT-HAm when cells were incubated in galactose (Figure 2A, 2), indicating moderate rereplication. We were concerned that this apparent rereplication signal could be spurious and due to drifts in the DNA flow cytometry signal in cells that enlarged due to cell-cycle arrest, even without actual rereplication. We observed, however, that Ddc2-GFP foci also accumulated in these cells (Figure 2B, 2, and 2C, 2), consistent with activation of rereplication (Archambault et al. 2005b; Green and Li 2005). Also, no apparent rereplication signal was detected using these methods with the clb1,3,4-del clb2-ts block, previously characterized as a G2 block without rereplication (Amon et al. 1993) (supplemental Figure S2 at http://www.genetics.org/supplemental/). For these reasons, we consider that this DNA flow cytometry signal likely represents authentic rereplication, but we recognize that in borderline cases it is hard to be sure of this. Recently, Green et al. (2006), using a microarray method, showed that rereplication at some specific loci could be detected even in cells without DNA flow-cytometry-detectable rereplication, and such an assay may ultimately be necessary to definitively prove rereplication in cases that are borderline by DNA flow cytometry.
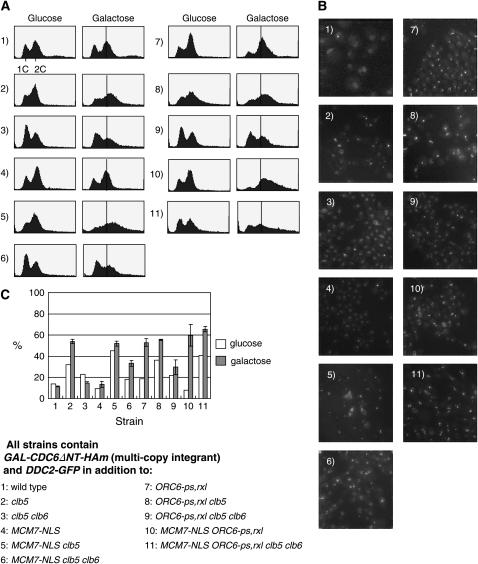
Figure 2.—
Induction of DNA rereplication and recruitment of Ddc2-GFP foci. (A) Cells were grown in YEP-D and transferred to YEP-R for 8 hr. Galactose at the final concentration of 3% was added for 4 hr. Cells were fixed and DNA content was measured by DNA flow cytometry analysis. The approximate position of 2C DNA content was determined on the basis of the major 2C peak in the control (1; GAL-CDC6ΔNT-HAm strain). This position is indicated by a bar. All samples were processed in parallel, and data were collected using the same set up (voltage 780 and total cell number 20,000 cells) and gating. As a rough empirical guide, we consider the DNA flow cytometry profile to represent over-replication if the main peak is shifted rightward compared to the bar. Each histogram is numbered on the basis of the strain numbering in Figure 1B. (B) The same strains, numbered as in A, were induced with galactose for 4 hr, and cells were observed under a DeltaVision microscope. Image stacks were deconvolved and processed at maximum projection. (C) Percentages of the cells showing DDC2-GFP foci in the presence of glucose or galactose. At least 100 cells were counted for each experiment, and three experiments were repeated. Average number and standard deviation were plotted. Open bars show percentages of DDC2-GFP foci when incubated in glucose, and shaded bars show the same when incubated in galactose.
These interpretations led to the idea that Clb5 blocks rereplication in cells with stabilized Cdc6. The DNA flow cytometry phenotype, as well as the Ddc2-GFP accumulation, was rescued by additional deletion of CLB6, implying that, in clb5 GAL-CDC6ΔNT-HAm cells, Clb6 induces inviability, rereplication, and DNA damage (Figure 1B; compare Figure 2A, 2 with 3; 2B, 2 with 3; and 2C, 2 with 3). This result implies that Clb6 can induce replication without blocking rereplication.
Clb5 blocks Clb6-dependent rereplication in cells containing a constitutively nuclear Mcm complex:
We tested the cyclin requirements for rereplication control when Mcm7 is constitutively localized in the nucleus. The MCM7-NLS mutation prevents cell-cycle-dependent Clb-Cdk regulation of Mcm complex localization. Mcm localization contributes to control of rereplication in parallel to Cdc6- and ORC-based mechanisms (Labib et al. 1999; Nguyen et al. 2000, 2001; Wilmes et al. 2004). GAL-CDC6ΔNT-HAm MCM7-NLS cells were viable on YEP-G, and these cells did not exhibit DNA rereplication or Ddc2-GFP foci (Figure 1B and Figure 2A, 4, 2B, 4, and 2C, 4), consistent with previous results (Nguyen et al. 2001). Deleting CLB5 in this background caused rereplication, DNA damage, and inviability (Figure 1B and Figure 2A, 5, 2B, 5, and 2C, 5). Consistent with the results above, additional deletion of CLB6 partially rescued inviability, largely prevented DNA rereplication, and reduced accumulation of Ddc2-GFP foci (Figure 1B, 2A, 6, 2B, 6, and 2C, 6). Inducing early accumulation of the mitotic cyclins Clb1–4 by deletion of CDH1 partially rescued the lethality in clb5 GAL-CDC6ΔNT-HAm MCM7-NLS and largely rescued the rereplication profile (Figure 3, A and B, top).
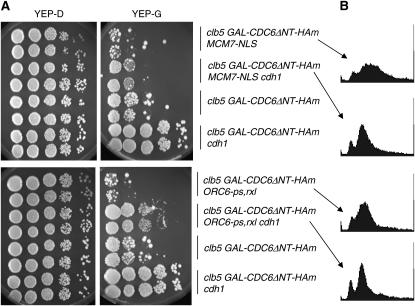
Figure 3.—
Cdh1 can promote inviability and rereplication in the absence of specific replication control mechanisms. (A) Serial dilutions as in Figure 1 for clb5 GAL-CDC6ΔNT-HAm ORC6-ps,rxl and clb5 GAL-CDC6ΔNT-HAm MCM7-NLS, with and without CDH1. (B) DNA flow cytometry analysis performed for selected strains after GAL-CDC6ΔNT-HAm induction, as in Figure 2.
Clb5 blocks Clb6-dependent rereplication in cells with unphosphorylatable Orc6 lacking a cyclin-binding motif:
We next examined the consequences of deleting S-phase cyclins in the presence of the ORC6-ps,rxl mutation, which eliminates cyclin binding and phosphorylation of Orc6 (Wilmes et al. 2004). Introducing ORC6-ps,rxl in GAL-CDC6ΔNT-HAm cells caused an accumulation of 2C DNA cells (Figure 2A, 7) and moderate slow growth (Figure 1B). Additionally deleting CLB5 in this background induced complete inviability (Figure 1B) and overt rereplication (Figure 2A, 8), indicating that Clb5 has Orc6-independent targets preventing rereplication. Potential additional targets for Clb5 include Orc2 phosphorylation and Mcm complex phosphorylation leading to Mcm nuclear export (see above). Lethality, rereplication, and DNA damage in this background were dependent on Clb6 (Figure 1B; compare Figure 2A, 8 with 9, and 2C, 8 with 9).
Cyclin requirements for rereplication in cells lacking both Mcm control and Orc control:
MCM7-NLS in a clb5 ORC6-ps,rxl background led to lethality in tetrad analysis even without overexpression of stabilized Cdc6 (data not shown); thus these strains could not be assayed directly for the rereplication phenotype, since in all other experiments we used conditional expression of stabilized Cdc6 to trigger rereplication and lethality. As in other contexts, however, additional deletion of CLB6 rescued the inviability of this background, in the absence but not in the presence of GAL-CDC6ΔNT expression (Figure 1B). These results broaden our findings to a context not requiring Cdc6 stabilization.
GAL-CDC6ΔNT-HAm MCM7-NLS ORC6-ps,rxl induced lethality, extensive rereplication, and multiple Ddc2-GFP foci per cell (Figure 1B; Figure 2A, 10, and 2B, 10), as reported previously (Nguyen et al. 2001; Wilmes et al. 2004; Archambault et al. 2005b; Green and Li 2005). Additional deletions of both CLB5 and CLB6 in this background did not rescue these phenotypes efficiently (Figure1B; Figure 2A, 11, 2B, 11, and 2C, 11), indicating that rescue by CLB5 and CLB6 deletions in other backgrounds was largely dependent on regulation of Cdc6, MCM, or ORC.
Clb6-dependent rereplication can be rescued by early accumulation of mitotic cyclins:
Above, we described three backgrounds with impaired control of rereplication in which Clb6 was deduced to cause rereplication, DNA damage foci, and loss of viability: clb5 GAL-CDC6ΔNT-HAm, clb5 GAL-CDC6ΔNT-HAm MCM7-NLS, and clb5 ORC6-ps,rxl GAL-CDC6ΔNT-HAm. In all of these backgrounds, additional deletion of CDH1 significantly rescued the phenotypes (Figures 1 and 3). Thus, we infer that the damaging rereplication induced by Clb6 in these backgrounds is suppressed by the simultaneous presence of mitotic cyclins along with Clb6, and we infer that the mitotic cyclins must therefore be able to effectively impose rereplication control by either the ORC-based or the Mcm-based mechanisms.
Stabilized Clb6 substitutes efficiently for Clb5 in regulating replication:
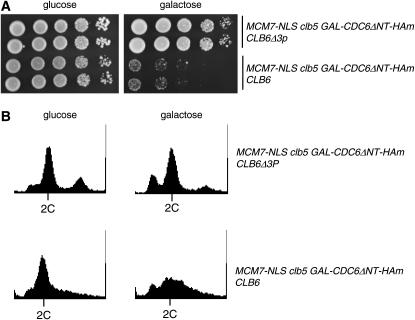
Clb5 and Clb6 degradation are regulated by distinct mechanisms (Jackson et al. 2006). Clb6p is mostly degraded at the G1/S transition through the SCFCdc4 ubiquitin ligase complex, while Clb5 is stable until mitosis and is degraded by APCCdc20. Mutation of three phosphorylation sites in Clb6 (CLB6Δ3P) leads to Clb6 stabilization by preventing SCFCdc4 binding (Jackson et al. 2006). As shown above, inviability and rereplication in the MCM7-NLS clb5 GAL-CDC6ΔNT-HAm background are strongly dependent on the absence of clb5 (Figures 1 and 2). We found that substitution of CLB6Δ3P for CLB6 in the MCM7-NLS clb5 GAL-CDC6ΔNT-HAm background completely rescued its strong lethality and also rescued its apparent rereplication defect as judged by DNA flow cytometry (Figure 4). Thus, Clb6 and Clb5 probably have similar intrinsic abilities to regulate rereplication, but Clb6 is normally unable to carry out this role due to its rapid degradation early in the cell cycle.
Figure 4.—
Stabilized Clb6 reverses loss of viability and replication control due to absence of Clb5. (A) Serial dilutions for MCM7-NLS clb5 GAL-CDC6ΔNT-HAm with CLB6Δ3P [gene replacement, encoding Clb6 stabilized due to mutation of phosphorylation sites (Jackson et al. 2006)] or CLB6-wt. (B) DNA content was measured by DNA flow cytometry analysis, as in Figure 2A. There is a minor peak at 4C DNA content in the CLB6Δ3P MCM7-NLS clb5 GAL-CDC6ΔNT-HAm strain. Microscopically, we did not detect significant clumping or division problems in this strain (data not shown). At present we do not know the explanation of this peak.
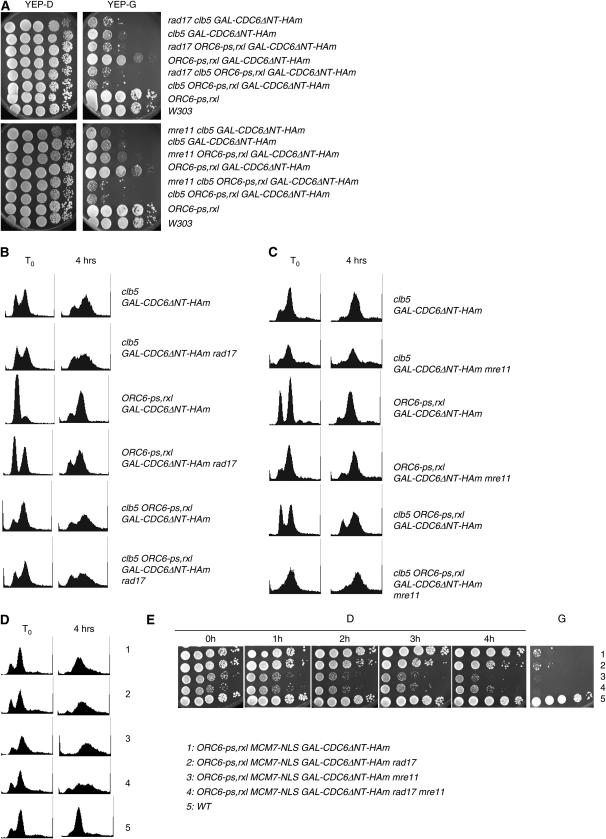
A DNA damage response including Mre11 and Rad17 functions to limit overt rereplication when cyclin-dependent mechanisms fail:
We previously showed that viability of ORC6-rxl GAL-CDC6ΔNTs cells depends on genes required for DNA damage response (Archambault et al. 2005b). Ddc2-GFP foci occurred in clb5 GAL-CDC6ΔNT-HAm and ORC6-ps,rxl GAL-CDC6ΔNT-HAm strains, suggesting that the DNA damage checkpoint might be activated (Figure 2B, 2 and 7, and 2C, 2 and 7). To test if the viability of clb5 GAL-CDC6ΔNT-HAm and ORC6-ps,rxl GAL-CDC6ΔNT-HAm depends on DNA damage signaling through the MRX complex or the 9-1-1 complex (Lowndes and Murguia 2000), cell viability was examined with or without MRE11 (MRX complex) or RAD17 (9-1-1 complex). The ORC6-ps,rxl GAL-CDC6ΔNT-HAm strain showed reduced viability in the absence of MRE11 or RAD17 (Figure 5A), suggesting that DNA damage occurs and is signaled or repaired by MRX and 9-1-1 complexes in those strains. Deleting MRE11 or RAD17 in the clb5 GAL-CDC6ΔNT-HAm had little effect, perhaps because the viability of clb5 GAL-CDC6ΔNT-HAm is already quite low. Deleting MRE11 or RAD17 had little effect on replication profiles in either background (Figure 5, B and C), suggesting that the increased inviability in the ORC6-ps,rxl GAL-CDC6ΔNT-HAm background caused by the MRE11 or RAD17 background is not caused by further deregulation of rereplication, but probably rather by response to DNA breaks generated by S-phase-cyclin-dependent rereplication.
Figure 5.—
Requirement for DNA damage response genes MRE11 and RAD17 in rereplicating cells. (A) Cell viability was tested in the presence or absence of MRE11 or RAD17 in cells of the indicated genotypes, as in Figure 1. (B–D) DNA content was analyzed by DNA flow cytometry after galactose induction as in Figure 2. (E) Cells were grown in YEP-D and transferred to YEP-R for 8 hr. Galactose at the final concentration of 3% was added. The samples were collected every hour after the galactose induction and were subjected to serial dilution experiments. The serial dilution was performed on YEP-D plates after the incubation in YEP-G media. As a control, cells were plated on YEP-G after the incubation in YEP-D media. Genotypes are as indicated.
In contrast, in the ORC6-ps,rxl MCM7-NLS GAL-CDC6ΔNT-HAm background (shown above to induce rereplication largely independent of S-phase cyclins), deleting MRE11 or RAD17 caused increased rereplication (Figure 5D), as reported previously (Archambault et al. 2005b).
The double deletion of MRE11 and RAD17 did not significantly enhance the rereplication seen in single deletion of either MRE11 or RAD17 (Figure 5D). We conclude that MRE11 and RAD17 are in largely overlapping pathways that can contribute to limit the extent of DNA rereplication when cyclin-Cdk-dependent rereplication blocks at the pre-RC level fail.
We assessed the ability of the MRX or 9-1-1 complexes to protect against the deleterious effects of loss of replication control by pulsing ORC6-ps,rxl MCM7-NLS GAL-CDC6ΔNT-HAm in galactose for short periods, followed by plating on glucose medium to assess viability. The MRE11 RAD17 controls showed little lethality after 4 hr in galactose medium, despite some amount of detectable overreplication (Figure 5E). Deletion of MRE11 in this background resulted in a 100-fold loss of viability upon short galactose pulses, while deletion of RAD17 resulted in at most a 10-fold viability loss. rad17 mre11 double mutants were not more sensitive than mre11 single mutants.
DISCUSSION
Cell-cycle regulators determine the viability of cells with compromised rereplication controls:
All of the mutant data presented in this article are summarized in Table 2, which emphasizes deductions that can be made from single additional deletions even in complex genetic backgrounds.
TABLE 2.
Summary of conclusions derived from genetic interactions and comparisons
| Strains and additional mutations (underlined) | Viability (serial dilution) | Rereplication (DNA flow cytometry analysis) |
|---|---|---|
| In CDC6ΔNT (no. 1) | Viable | No rereplication |
| clb5 from no. 2 | Clb5 stays alive | Clb5 blocks rereplication |
| MCM7-NLS from no. 4 | Mcm regulation has no effect | Mcm regulation has no effect |
| ORC6-ps,rxl from no. 7 | Orc regulation helps to stay alive | Orc regulation has no effect |
| In CDC6ΔNT clb5 (no. 2) | Reduced viability | Moderate rereplication |
| clb6 from no. 3 | Clb6 kills | Clb6 may drive rereplication |
| MCM7-NLS from no. 5 | Mcm regulation helps viability | Mcm regulation blocks rereplication |
| ORC6-ps,rxl from no. 8 | Orc regulation helps viability | Orc regulation may block rereplication |
| cdh1 from Figure 1 | Cdh1 kills | No data |
| In ORC6-ps,rxl CDC6ΔNT (no. 7) | Moderate reduced viability | No rereplication (2C DNA accumulation) |
| clb5 from no. 8 | Clb5 stays alive | Clb5 blocks rereplication |
| MCM7-NLS from no. 10 | Mcm regulation stays alive | Mcm regulation blocks rereplication |
| sic1 from Figure 1 | Sic1 regulation stays alive | No data |
| In ORC6-ps,rxl clb5 CDC6ΔNT (no. 8) | Lethal | Rereplication |
| clb6 from no. 9 | Clb6 kills | Clb6 drives rereplication |
| MCM7-NLS from tetrad | Mcm regulation stays alive | Mcm cannot test |
| cdh1 from Figure 3 | Cdh1 may kill | Cdh1 drives rereplication |
| In MCM7-NLS CDC6ΔNT (no. 4) | Viable | No rereplication |
| clb5 from no. 5 | Clb5 stays alive | Clb5 blocks rereplication |
| ORC6-ps,rxl from no. 10 | Orc regulation stays alive | Orc regulation blocks rereplication |
| In MCM7-NLS clb5 CDC6ΔNT (no. 5) | Lethal | Rereplication |
| clb6 From #6 | Clb6 helps killing | Clb6 drives rereplication |
| CLB6Δ3p from Figure 4 | Clb6 kills because of early proteolysis | Clb6 cannot block rereplication because of early proteolysis |
| cdh1 from Figure 3 | Cdh1 helps to kill | Cdh1 drives rereplication |
| In ORC6-ps,rxl MCM7-NLS CDC6ΔNT (no. 10) | Lethal | Rereplication |
| clb5 clb6 from no. 11 | Clb5,6 have little effect | Clb5,6 have little effect |
| In ORC6-ps,rxl MCM7-NLS (no. 10 on YEP-D) | Viable | No rereplication |
| clb5 from tetrad | Clb5 stays alive | Clb5 cannot be tested |
| In ORC6-ps,rxl MCM7-NLS clb5 | Lethal | Cannot test |
| clb6 from no. 11 on YEP-D | Clb6 kills | Clb6 cannot be tested |
| In clb5 clb6 GAL-CDC6ΔNT (no. 3) | Viable | No rereplication |
| MCM7-NLS from no. 6 | Mcm regulation may stay alive | Mcm regulation has no effect |
| ORC6-ps,rxl from no. 9 | Orc regulation has no effect | Orc regulation has no effect |
All of the mutant strains presented in this study are summarized. Strains that are underlined were compared to those with a single additional mutation. Results from tetrad analysis, serial dilution (Figures 1B, 3, and 4), and rereplication analysis (Figure 2A) are considered. “no.” refers to the strain from Figure 1B and Figure 2. “Orc regulation” refers to the ability to bind to and/or phosphorylate Orc6 and is based on the effects of the ORC6-ps,rxl mutation. “Mcm regulation” refers to the ability to regulate nuclear localization of the Mcm complex and is based on the effects of the MCM7-NLS mutation. See text.
Several controls are known to function in preventing rereplication in S. cerevisiae. Cdk-dependent phosphorylation of Cdc6 on its N terminus promotes ubiquitination and degradation (Drury et al. 1997, 2000; Nguyen et al. 2001) and Cdc6 sequestration by Clb2-Cdk1 (Mimura et al. 2004); Cdk-dependent phosphorylation of Orc6 and Orc2 is thought to hinder pre-RC formation (Nguyen et al. 2001); and Cdk-dependent phosphorylation of MCM proteins causes cytoplasmic retention of the complex (Labib et al. 1999; Nguyen et al. 2000, 2001; Liku et al. 2005). In addition, binding of Clb5 to Orc6 via a defined RXL motif contributes to preventing rereplication (Wilmes et al. 2004).
Our results support the idea that the timing of Clb5 and Clb6 activity relative to other cyclins is crucial in determining the viability of rereplication-sensitized strains, in which Cdc6 is stabilized (CDC6ΔNT) and Clb5-dependent replication control mechanisms are disrupted (ORC6-rxl or clb5). In such strains, Clb6 may activate an additional round of replication from some reloaded origins, since Clb1-4, which can efficiently limit rereplication even in these partially compromised strains, only accumulate later [their accumulation may be additionally delayed in clb5 mutants due to prolonged Cdh1 activity (Yeong et al. 2001)]. In this interpretation, the earlier that Clb5,6 activity appears relative to mitotic cyclins in cells lacking Clb5-dependent mechanisms of control, the more damage is likely to be done. This could account for the enhanced lethality caused by deletions of SIC1. Inversely, the less time there is between Clb5,6 activity onset and mitotic cyclin accumulation, the less damage is done. This could account for the rescue caused by simultaneous deletion of CLB5 and CLB6 (in which case both initiation and regulation of rereplication can be carried out by Clb1-4) and by deletion of CDH1 or by replacement of clb5 with CLB5pCLB2 (in these cases, the putative unregulated replication initiation driven by Clb6 is temporally accompanied by accumulation of some of Clb1-4, which can block reinitiation). This interpretation also implies that origin reloading must be blocked throughout S-phase, not merely in the extended G2 postreplicative period; this was the biological rationale that we proposed previously for efficient Clb5 binding locally to fired origins in mid-S-phase, even while other neighboring origins remained unfired (Wilmes et al. 2004). Recent results with a very different approach, genomewide mapping of DNA synthesis (Green et al. 2006), independently support the conclusion that control of origin reloading must function throughout S-phase.
Our results suggest that while Clb6 is active in promoting replication initiation, at least at some origins (Schwob and Nasmyth 1993; Donaldson et al. 1998), it is apparently much less able to block rereplication, due to its early, rapid proteolysis by SCFCdc4 ubiquitin ligase (Jackson et al. 2006) (Figure 4).
Previously, we concluded that the mechanism by which Clb5 blocks rereplication requires binding specifically to the regulated origin (Wilmes et al. 2004). There is not a large excess of Clb5 over the probable number of replication origins, and Clb6 is present at only ∼10% the level of Clb5 (Cross et al. 2002), presumably due to its early proteolysis (Jackson et al. 2006); therefore, Clb6 may simply not be present at a high-enough level to stoichiometrically inhibit origin reloading. The mechanism of promotion of origin firing, in contrast, probably does not involve stoichiometric Clb5,6 origin binding (Wilmes et al. 2004) and may instead rely on catalytic phosphorylation of trans-acting replication factors such as Sld2 (Masumoto et al. 2002), allowing even low levels of Clb6, which escape SCFCdc4-dependent proteolysis later in S-phase, to positively promote rereplication from reloaded origins.
Multiple levels of control of replication reinitiation:
In early S-phase, Clb5 and Clb6 initiate replication, and Clb5 simultaneously inhibits rereplication, probably acting through phosphorylation of Mcm and the Orc complex, since both are likely to be favored phosphorylation targets of Clb5 (Wilmes et al. 2004; Archambault et al. 2005a; Loog and Morgan 2005).
A clb1 clb2(ts) clb3 clb4 strain (Amon et al. 1993) additionally containing ORC6-ps,rxl GAL-CDC6ΔNT(m) showed G2 accumulation and no rereplication at a nonpermissive temperature in galactose (supplemental Figure S2 at http://www.genetics.org/supplemental/). In this strain, replication is driven exclusively by Clb5,6 (Schwob et al. 1994). This supports the hypothesis that S-phase cyclins, in addition to targeting Orc6, can regulate other rereplication-limiting substrates independently of Orc6 binding or Orc6 phosphorylation, such as the Mcm complex (Loog and Morgan 2005). In the converse situation, when initiation is driven in the absence of CLB5,6 and all replication is under control of Clb1-4 (Schwob et al. 1994), mitotic cyclins inhibit rereplication, presumably also through largely redundant regulation of Mcm and Orc (Figure 2; compare strains 3, 6, 9, and 11). Thus, there is redundancy among the B-type cyclins, and additionally among the multiple phosphorylation targets Cdc6, Orc2, Orc6, and the Mcm complex, and many of these cyclins and/or controls can be eliminated without strong induction of rereplication.
While our results clearly imply that different B-type cyclins are substantially redundant for rereplication control by multiple mechanisms, we were interested in the possibility of some degree of specificity. One result suggestive of specificity was the finding that MCM7-NLS GAL-CDC6ΔNT cells exhibited normal growth rates, but deletion of CLB5,6 in this background caused reduced viability (compare strains 4 and 6 in Figure 1B); in sharp contrast, ORC6-ps,rxl GAL-CDC6ΔNT cells exhibit slow growth, and deletion of CLB5,6 resulted in complete rescue to normal growth rates (compare strains 7 and 9 in Figure 1B). The opposite effects of CLB5,6 deletion in these backgrounds could be explained if replication under control of Clb5,6 specifically involved rereplication control via ORC, while replication under control of Clb1-4 specifically involved rereplication control via MCM. However, the fact that these viability phenomena were not reflected in DNA flow-cytometry-detectable rereplication (Figure 2A) reduces our ability to simply interpret these results. As discussed above, the results with DNA flow-cytometry-detectable rereplication are most consistent with all B-type cyclins (except Clb6, due to its rapid proteolysis) having the ability to regulate rereplication via either ORC or MCM.
Finally, we showed that viability of ORC6-ps,rxl CDC6ΔNT-HAm relies on the DNA damage checkpoint genes RAD17 and MRE11. However, the rereplication profile by DNA flow cytometry analysis in this background did not change when RAD17 or MRE11 was removed (Figure 5, B and C). This suggests that inhibition of rereplication by mitotic Clb1-4 kinases, and not the DNA damage response, is the primary mechanism preventing rereplication when Clb5-dependent mechanisms are disrupted. On the other hand, removing DNA damage signaling in MCM7-NLS ORC6-ps,rxl CDC6ΔNT-HAm enhanced rereplication (Figure 5D). Therefore, it appears that restraint of rereplication by a DNA-damage response pathway acts as a “last resort” mechanism when cyclin-CDK-dependent mechanisms fail.
Sensitive assays of the extent of rereplication of specific genomic regions under conditions of complete or partial deregulation of replication control have indicated that additional levels of control exist. Not all origins are equally used for rereplication, and some are used multiple times (Green et al. 2006; Tanny et al. 2006); even with only a few replication controls removed, where no detectable rereplication exists by DNA flow cytometry, rereplication of some regions can still be detected (Green et al. 2006). The latter result emphasizes the likely evolutionary reason for the apparent multiplicity of controls; it is unknown if different regions differ in their cyclin requirements for regulation of rereplication. Many origins are not used detectably for rereplication; additionally, it was shown that origin reloading with the Mcm complex may be necessary but not sufficient for overt rereplication (Tanny et al. 2006). Thus, there are probably additional controls blocking rereplication that remain to be identified.
Acknowledgments
We thank Steven Haase for the CLB6Δ3P strain. Funding was provided by PHS grant GM047238 to F.R.C. A.E.I. was supported by National Institutes of Health training grant T32 CA09673.
References
- Amon, A., M. Tyers, B. Futcher and K. Nasmyth, 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74: 993–1007. [DOI] [PubMed] [Google Scholar]
- Aparicio, O. M., D. M. Weinstein and S. P. Bell, 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69. [DOI] [PubMed] [Google Scholar]
- Archambault, V., E. J. Chang, B. J. Drapkin, F. R. Cross, B. T. Chait et al., 2004. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14: 699–711. [DOI] [PubMed] [Google Scholar]
- Archambault, V., N. E. Buchler, G. M. Wilmes, M. D. Jacobson and F. R. Cross, 2005. a Two-faced cyclins with eyes on the targets. Cell Cycle 4: 125–130. [DOI] [PubMed] [Google Scholar]
- Archambault, V., A. E. Ikui, B. J. Drapkin and F. R. Cross, 2005. b Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 25: 6707–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S. P., and A. Dutta, 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333–374. [DOI] [PubMed] [Google Scholar]
- Broek, D., R. Bartlett, K. Crawford and P. Nurse, 1991. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature 349: 388–393. [DOI] [PubMed] [Google Scholar]
- Calzada, A., M. Sanchez, E. Sanchez and A. Bueno, 2000. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem. 275: 9734–9741. [DOI] [PubMed] [Google Scholar]
- Cocker, J. H., S. Piatti, C. Santocanale, K. Nasmyth and J. F. Diffley, 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379: 180–182. [DOI] [PubMed] [Google Scholar]
- Cross, F. R., M. Yuste-Rojas, S. Gray and M. D. Jacobson, 1999. Specialization and targeting of B-type cyclins. Mol. Cell 4: 11–19. [DOI] [PubMed] [Google Scholar]
- Cross, F. R., V. Archambault, M. Miller and M. Klovstad, 2002. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell 13: 52–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann, C., J. F. Diffley and K. A. Nasmyth, 1995. S-phase-promoting cyclin-dependent kinases prevent rereplication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 5: 1257–1269. [DOI] [PubMed] [Google Scholar]
- Devault, A., E. A. Vallen, T. Yuan, S. Green, A. Bensimon et al., 2002. Identification of Tah11/Sid2 as the ortholog of the replication licensing factor Cdt1 in Saccharomyces cerevisiae. Curr. Biol. 12: 689–694. [DOI] [PubMed] [Google Scholar]
- Diffley, J. F., J. H. Cocker, S. J. Dowell and A. Rowley, 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303–316. [DOI] [PubMed] [Google Scholar]
- Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer et al., 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2: 173–182. [DOI] [PubMed] [Google Scholar]
- Drury, L. S., G. Perkins and J. F. Diffley, 1997. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16: 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury, L. S., G. Perkins and J. F. Diffley, 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10: 231–240. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Y. Chi, P. Yang and J. L. Campbell, 1999. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10: 3263–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, C. B., and F. R. Cross, 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6: 1695–1706. [DOI] [PubMed] [Google Scholar]
- Green, B. M., and J. J. Li, 2005. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 16: 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B. M., R. J. Morreale, B. Ozaydin, J. L. Derisi and J. J. Li, 2006. Genomewide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol. Biol. Cell 17: 2401–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles, J., D. Fisher, A. Woollard and P. Nurse, 1994. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78: 813–822. [DOI] [PubMed] [Google Scholar]
- Jackson, L. P., S. I. Reed and S. B. Haase, 2006. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol. Cell. Biol. 26: 2456–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey, S. E., and S. Cotterill, 2003. Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol. Cell 12: 1067–1075. [DOI] [PubMed] [Google Scholar]
- Labib, K., J. F. Diffley and S. E. Kearsey, 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1: 415–422. [DOI] [PubMed] [Google Scholar]
- Liku, M. E., V. Q. Nguyen, A. W. Rosales, K. Irie and J. J. Li, 2005. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol. Biol. Cell 16: 5026–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog, M., and D. O. Morgan, 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434: 104–108. [DOI] [PubMed] [Google Scholar]
- Lowndes, N. F., and J. R. Murguia, 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10: 17–25. [DOI] [PubMed] [Google Scholar]
- Luo, K. Q., S. Elsasser, D. C. Chang and J. L. Campbell, 2003. Regulation of the localization and stability of Cdc6 in living yeast cells. Biochem. Biophys. Res. Commun. 306: 851–859. [DOI] [PubMed] [Google Scholar]
- Masumoto, H., S. Muramatsu, Y. Kamimura and H. Araki, 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655. [DOI] [PubMed] [Google Scholar]
- Mendenhall, M. D., 1993. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science 259: 216–219. [DOI] [PubMed] [Google Scholar]
- Mimura, S., T. Seki, S. Tanaka and J. F. Diffley, 2004. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 431: 1118–1123. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., C. Co, K. Irie and J. J. Li, 2000. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr. Biol. 10: 195–205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., C. Co and J. J. Li, 2001. Cyclin-dependent kinases prevent DNA rereplication through multiple mechanisms. Nature 411: 1068–1073. [DOI] [PubMed] [Google Scholar]
- Schwab, M., M. Neutzner, D. Mocker and W. Seufert, 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20: 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, E., and K. Nasmyth, 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7: 1160–1175. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244. [DOI] [PubMed] [Google Scholar]
- Tanaka, S., and J. F. Diffley, 2002. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nat. Cell Biol. 4: 198–207. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., D. Knapp and K. Nasmyth, 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649–660. [DOI] [PubMed] [Google Scholar]
- Tanny, R. E., D. M. Macalpine, H. G. Blitzblau and S. P. Bell, 2006. Genomewide analysis of rereplication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell 17: 2415–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., S. Prinz and A. Amon, 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463. [DOI] [PubMed] [Google Scholar]
- Wasch, R., and F. R. Cross, 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418: 556–562. [DOI] [PubMed] [Google Scholar]
- Wilmes, G. M., V. Archambault, R. J. Austin, M. D. Jacobson, S. P. Bell et al., 2004. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 18: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong, F. M., H. H. Lim, Y. Wang and U. Surana, 2001. Early expressed Clb proteins allow accumulation of mitotic cyclin by inactivating proteolytic machinery during S phase. Mol. Cell. Biol. 21: 5071–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]