Abstract
Fragile X mental retardation proteins (FMRP) are RNA-binding proteins that interact with a subset of cellular RNAs. Several RNA-binding domains have been identified in FMRP, but the contribution of these individual domains to FMRP function in an animal model is not well understood. In this study, we have generated flies with point mutations in the KH domains of the Drosophila melanogaster fragile X gene (dfmr1) in the context of a genomic rescue fragment. The substitutions of conserved isoleucine residues within the KH domains with asparagine are thought to impair binding of RNA substrates and perhaps the ability of FMRP to assemble into mRNP complexes. The mutants were analyzed for defects in development and behavior that are associated with deletion null alleles of dfmr1. We find that these KH domain mutations result in partial loss of function or no significant loss of function for the phenotypes assayed. The phenotypes resulting from these KH domain mutants imply that the capacities of the mutant proteins to bind RNA and form functional mRNP complexes are not wholly disrupted and are consistent with biochemical models suggesting that RNA-binding domains of FMRP can function independently.
THE fragile X mental retardation protein (FMRP) is an RNA-binding protein necessary for normal neuronal development and behavior in all species where its function has been examined. A general model for FMRP function is that it regulates nucleocytoplasmic transport, subcellular localization, and translation of select RNA transcripts (reviewed by Bardoni and Mandel 2002; Jin and Warren 2003; Jin et al. 2004a; Bagni and Greenough 2005). Biochemical analyses have uncovered several RNA-binding motifs associated with FMRP function, including two KH domains (hnRNP-K homology) and an arginine and glycine-rich motif (RGG box) that are common to RNA-binding proteins (Ashley et al. 1993; Siomi et al. 1993). The highly conserved N termini of FMRPs have RNA-binding capacity as well (Adinolfi et al. 1999, 2003). The N-terminal 110 amino acids of FMRPs are similar to Tudor/Agenet domains and are members of an extended family that is referred to as the Tudor domain “royal family” (Maurer-Stroh et al. 2003). This domain family is related to methyl-substrate-binding proteins that are implicated in regulation of chromatin structure and includes the chromodomain.
RNA substrates for FMRP have conserved elements in primary sequence and/or higher-order structures that interact with the aforementioned RNA-binding domains. A G-quartet structure within RNA interacts with the RGG box (Darnell et al. 2001; Schaeffer et al. 2001), and the second KH domain recognizes a loop–loop pseudoknot RNA structure referred to as a kissing complex (Darnell et al. 2005). A stem-loop structure within BC1 RNA is reported to interact specifically with the N-terminal 217 amino acids of FMRP (Zalfa et al. 2005; but see Wang et al. 2005 for an opposing view). These studies demonstrate that individual RNA-binding domains of FMRP have distinct substrates with which they interact and that the ability of these domains to bind substrates is not dependent upon other FMRP RNA-binding domains. Loss of function of any of these domains might then result in only a subset of RNA substrates losing the ability to bind FMRP.
Expansion of a CGG trinucleotide repeat in the 5′-UTR of the FMR1 gene, followed by methylation and transcriptional silencing, is the basis for the vast majority of fragile X cases in humans (see O'Donnell and Warren 2002 for a review of human fragile X inheritance patterns). Other alleles result from deletions or nonsense codons, and thus little structure–function information has been obtained from analysis of human FMR1 alleles. One significant exception is the substitution of a highly conserved isoleucine residue in KH domains to asparagine (I304N) within the second KH domain of human FMRP that is associated with unusually severe fragile X phenotypes (De Boulle et al. 1993). Until recently, models to explain the effects of the I304N substitution have been enigmatic. Defects in RNA binding have been proposed on the basis of the finding that the I304N protein is impaired in binding RNA homopolymers under high salt concentrations (Siomi et al. 1994) and the analysis of a co-crystal structure of a KH domain and RNA substrate (Lewis et al. 2000). Although the I304N protein can bind bulk poly(A) RNA (Feng et al. 1997), in contrast to wild-type FMRP, it does not associate with polyribosomes (Feng et al. 1997) or with itself (Laggerbauer et al. 2001). These results suggest that the inability to form proper messenger ribonucleoprotein (mRNP) complexes is a significant factor contributing to the I304N phenotype and have prompted suggestions that the severity of phenotypes associated with the mutation arise from dominant negative or antimorphic effects (Feng et al. 1997). The above biochemical studies have been reconciled by the findings that the second KH domain of FMRP binds kissing complex RNAs, the I304N substitution abolishes this association, and the kissing complex RNAs can compete FMRP off polyribosomes (Darnell et al. 2005). These results imply that FMRP association with polyribosomes is dependent upon an interaction of the second KH domain with RNAs containing a kissing complex structure.
Many studies demonstrate that the Drosophila fragile X protein shares biochemical functions with its vertebrate counterparts and regulates similar neural and behavioral functions (reviewed by Gao 2002; Jin and Warren 2003; Dölen and Bear 2005; Zhang and Broadie 2005). Previously existing alleles of dfmr1 are strong or null alleles resulting from imprecise P-element excisions or nonsense codons (Zhang et al. 2001; Dockendorff et al. 2002; Lee et al. 2003). Although KH domain mutations exist for dfmr1 as cDNA constructs, the misexpression or overexpression of these alleles in neural and muscle tissues via the GAL4-UAS system results in physiologic abnormalities or cell death (Wan et al. 2000; Zhang et al. 2001). Starting with a genomic rescue fragment encompassing dfmr1, we have created derivatives of this rescue fragment where conserved isoleucine residues in the KH domains have been mutated as a means to assess the importance of these domains in an animal model. These P-element-borne transgenes have been recombined onto a chromosome deleted for dfmr1 to produce animals that express only mutant forms of dFMR1 protein. Our analyses of flies with the KH domain mutations show that they result in either partial or no loss of function of the behavior and developmental phenotypes that were examined. These findings show that the mutant proteins retain a significant degree of function in vivo and are consistent with biochemical models that predict FMRP RNA-binding domains as having some independent functions. These alleles of dfmr1 will be useful tools for both genetic and biochemical screens in identifying RNAs and proteins that interact with the fragile X protein.
MATERIALS AND METHODS
Generation of KH domain mutants:
A subclone of a 14-kb BamHI–StuI genomic fragment spanning the dfmr1 locus was subjected to site-directed mutagenesis via the mega-primer technique, using a proofreading polymerase (methods compiled in Sambrook and Russell 2001). All PCR-amplified fragments were sequenced to confirm the presence of the desired mutation and the absence of secondary mutations resulting from base misincorporations during amplification. Mutant DNA was then substituted for the corresponding wild-type fragment and the resulting mutant rescue fragments were cloned into pCaSpeR-4 (Pirrotta 1988) for subsequent transformation. Transformations were done under conditions described in Spradling and Rubin (1982). We found the transformation efficiency to be very low, with one transformed fly appearing in ∼300 G0 crosses for each of the mutant transgenes. Both mutant transgenes mapped to the third chromosome, and thus the transgene insertions were recombined onto a chromosome harboring the dfmr13 allele, which removes the dfmr1 open reading frame (Dockendorff et al. 2002; Pan et al. 2004). These transgenes were judged to map within two recombination units of the endogenous dfmr1 locus on the basis of the frequency with which recombination of the two loci occurred. The resulting stocks are of the following genotypes: P[dfmr1I244N]w+ dfmr13/TM6C Tb Sb and P[dfmr1I307N]w+ dfmr13/TM6C Tb Sb. These recombinant stocks were then crossed to flies with the dfmr13 allele to produce animals heterozygous for the transgene insertion and homozygous for the dfmr13 allele. Thus, the only dFMR1 protein produced in such animals is from the mutant allele. For clarity, throughout the text and figures these stocks will be referred to simply by the nature of the KH domain substitution. To test for effects of an increased dosage, we crossed the stocks with the transgenes recombined onto the dfmr13 null chromosome to a stock expressing the Δ2-3 transposase. Flies were selected that had enhanced expression of the mini-white marker and of mutant dFMR1 protein, indicative of a replicative transposition event. For both transgenes, the second copy of the insertion mapped to the third chromosome, and these stocks were balanced using TM6C Tb Sb. To differentiate these stocks from those with the single copy of the transgene, we refer to them in figures and text with the suffix “2X.”
Fly stocks, genetics, and culture:
All dfmr1 mutant stocks in this study were derived from a w1118 background and maintained on a yeast–cornmeal–molasses medium at 25°. The third chromosome balancer TM6C Tb Sb was used to maintain dfmr1 alleles.
Courtship and circadian behavior analyses:
For courtship behavior testing, males of the appropriate genotypes were collected within 2 hr of eclosion and kept in isolation prior to testing. Female targets were of the genotype XX, y, f (attached X) and collected as virgins for courtship testing. All flies were kept in 12:12 light/dark (LD) cycles at 25° and 70–75% relative humidity and were aged for 4 days prior to analysis. For the naive courtship analysis, the 4-day-old male and female were transferred via aspiration to a mating chamber 20 mm in diameter and 5 mm deep. These chambers were kept in humidified conditions throughout the assay. Transferred males were given a 5-min recovery period prior to addition of the female target. All assays were performed within 30 min of the change in light cycle. Males were monitored for courtship activity that included following of the female, wing extension and vibration, tapping of the female with his foreleg, and attempted copulation for a period of 10 min or until copulation occurred. The percentage of time that the male spent in active pursuit of the female was recorded as the courtship index. A minimum of 25 animals was tested for each genotype.
Circadian behavior was tested as described in Dockendorff et al. (2002). Flies were entrained to a 12:12 light/dark cycle, placed into activity monitors (Trikinetics, Waltham, MA), maintained in light/dark cycles, and then placed under constant darkness. Locomotion activity was collected in 30-min bins. The percentage of flies judged to be rhythmic was assessed by Clocklab software (Actimetrics, Evanston, IL) as follows: Using a confidence level of 0.025, batch analyses were performed for the genotypes tested, monitoring the locomotion activity in constant darkness over 7 days. The difference between the power (1) and significance (1) values was calculated for each fly, and a value of <10 was the basis for judging an arrhythmic phenotype. Visual analyses of periodograms and actograms were also conducted to confirm the results.
Antibodies and immunocytochemistry:
Larval neuromuscular junction (NMJ) type I boutons were detected by staining third instar larval fillets with antihorseradish peroxidase (Cappel, Aurora, OH) at a dilution of 1:200. Mushroom bodies were visualized by staining whole mounts of brains with anti-FasII at a 1:10 dilution (mAb 1D4 obtained from University of Iowa Developmental Studies Hybridoma Bank). Secondary antibodies conjugated to either HRP or fluorochrome were obtained from Jackson ImmunoResearch (West Grove, PA) and used at a 1:200 dilution. Confocal images were collected on an Olympus FV500 microscope. Western blots were performed as described in Wan et al. (2000) using anti-dFMR1 antibody 5A11 at a 1:1000 dilution and anti-β-tubulin mAb E7 (both from the University of Iowa Developmental Studies Hybridoma Bank) at a 1:400 dilution.
Statistical analyses:
Courtship indexes were arcsin transformed and then analyzed by one-way ANOVA, followed by a Tukey–Kramer post-test. NMJ bouton counts were analyzed by one-way ANOVA with a Tukey–Kramer post-test or by a Kruskal–Wallis test, followed by a Dunn post-test. The analyses of courtship indexes and NMJ bouton counts were conducted using InStat software from GraphPad (San Diego). Comparisons of mushroom-body (MB) axon midline crossings and circadian rhythmicity were made by a chi-square test for homogeneity.
RESULTS
Generation of point mutations in KH domains of dfmr1:
A 14-kb genomic rescue fragment has previously been shown to rescue all known behavioral and developmental phenotypes associated with dfmr1 loss of function(Dockendorff et al. 2002; Lee et al. 2003; Michel et al. 2004; Costa et al. 2005). Subclones of this fragment were subjected to site-directed mutagenesis to convert highly conserved isoleucine residues in the two KH domains to asparagines (I244N, I307N for the Drosophila protein; Figure 1A). These substitutions are predicted to strongly inhibit binding of cognate RNA substrates (Lewis et al. 2000; Darnell et al. 2005) and may interfere with folding of the KH domain (Musco et al. 1996, 1997). Upon reconstruction of mutated sequences to the rescue fragment, the mutant rescue fragments were introduced to flies via P-element transformation and then recombined to chromosomes harboring the dfmr13 allele, a deletion null allele where the entire open reading frame of dfmr1 is removed (Dockendorff et al. 2002; Pan et al. 2004). Crossing such chromosomes to the dfmr13 chromosome results in flies heterozygous for the P-element transgenes and that express only the mutant alleles under control of the endogenous dfmr1 promoter. Since it might be expected that an I244N I307N double mutant could have an additive effect on any phenotypes observed with the single KH domain mutants, we attempted to obtain stocks with such an allele. Despite extensive efforts, we failed to obtain transgenic animals that harbored an I244N I307N double mutation of dfmr1 that was expressed via its endogenous promoter even in an otherwise wild-type background. Although UAS-GAL4 overexpression of a dfmr1 cDNA with the I244N I307N double mutation in the developing eye fails to induce a rough eye phenotype (Wan et al. 2000), we have observed that overexpression of the same transgene by myosin heavy chain-GAL4 can be lethal to pupae, indicating that such an allele can have dominant deleterious effects in specific tissues (T. C. Dockendorff, unpublished observations).
Figure 1.—
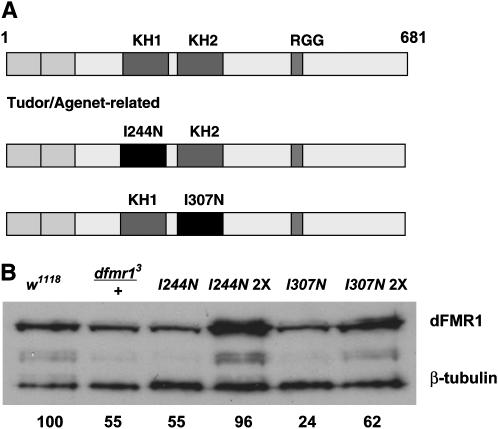
Schematic of fragile X protein and expression analysis of dfmr1 KH domain alleles. (A) RNA-binding domains of FMRP. Two KH domains, an RGG box, and two tandem copies of a Tudor/Agenet-related domain have all been demonstrated to bind RNA. KH domains with conserved isoleucine residues mutated to asparagine for this study are depicted, (B) Western blot of total fly extracts from wild-type, dfmr13 heterozygote, and flies expressing one or two (2X) copies of a transgene harboring a dfmr1 genomic rescue fragment coding for either an I244N or an I307N substitution in the KH domains. Extracts were prepared from males aged 2–3 days. Signals were scanned and quantified by ImageQuant software (Molecular Dynamics, Sunnyvale, CA), and average expression levels compared to a w1118 control from six independent blots are given.
To discern the level of dFMR1 protein expression from the transgenes, Western blotting was performed on male flies harboring the mutant KH domain transgenes as the sole allele of dfmr1. These studies show that flies with a single copy of the transgene with the I244N allele express the mutant protein at a level very similar to that seen with a dfmr1 heterozygote. The single copy of the I307N transgene had ∼25% the expression of dFMR1 protein present in a control w1118 background (Figure 1B). To create stocks with increased doses of each transgene, the P elements were mobilized and stocks that had undergone replicative transposition resulting in elevated levels of the mutant proteins were selected. These stocks were judged to express mutant proteins at 97 and 62% of the level seen with w1118 for the I244N and I307N substitutions, respectively (Figure 1B). To control for dosage effects, dfmr1 heterozygotes are included in all of the following analyses. Since the human I304N substitution has been hypothesized to exert dominant effects, we examined our KH domain allele stocks for dominant phenotypes through analysis of flies that expressed both a mutant and wild-type allele of dfmr1. These stocks are noted in figures with both the KH domain allele designation and a “+” for the wild-type allele.
The NMJ bouton overgrowth phenotype associated with null alleles of dfmr1 is not observed with the I244N or I307N KH domain substitutions:
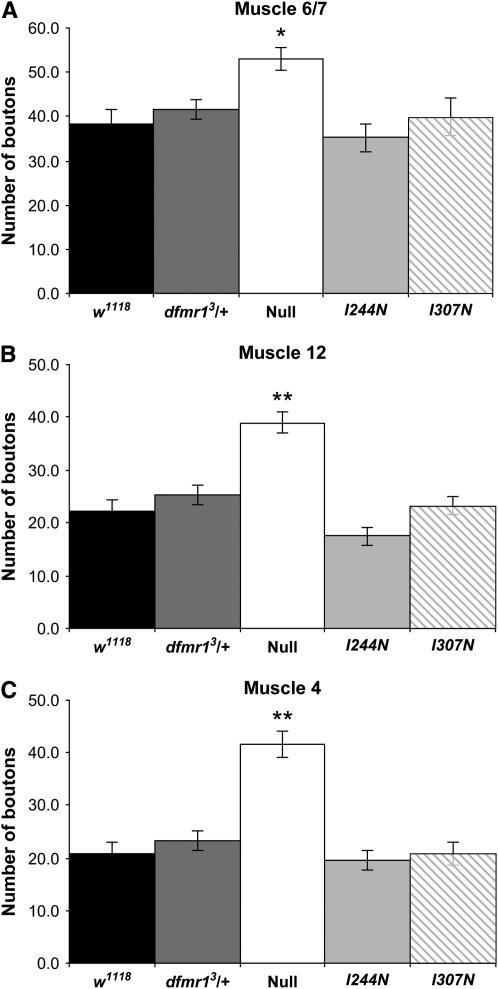
Strong or null alleles of dfmr1 result in an overgrowth of larval neuromuscular junction boutons (Zhang et al. 2001; Jin et al. 2004b). To assess the impact of the KH domain mutations on this phenotype, we analyzed the numbers of type I boutons at larval NMJs using flies with wild-type and null alleles of dfmr1 as controls. We examined muscles 4, 6/7, and 12 from segment A3 for the analysis of larval NMJs. Figure 2 shows that, in all muscles examined, there is no significant increase in bouton number over wild type or dfmr1 heterozygote controls from larvae with a single copy of a transgene expressing either I244N or I307N substitutions as the sole source of dFMR1 protein, while there is the expected pronounced overgrowth of boutons from larvae homozygous for the null allele of dfmr1. These results indicate that neither of the Ile → Asn substitutions in the KH domains affects the ability of dFMR1 to regulate larval NMJ bouton numbers and thus suggests that other domains of FMRP play a more vital role in this process.
Figure 2.—
Numbers of larval NMJ boutons are not increased in flies expressing KH domain I244N or I307N substitutions as a sole source of dFMR1 protein. Third instar larvae were dissected and probed with antibodies against horseradish peroxidase to assess numbers of type I NMJ boutons, which are increased in flies homozygous for strong or null alleles of dfmr1 (Zhang et al. 2001; Jin et al. 2004b). Analyses of several muscle types from segment A3 show that type I boutons are significantly increased in all muscle types examined from animals homozygous for a null allele of dfmr1 compared to all wild-type and KH domain alleles examined (P < 0.001 for muscle 4, Kruskal–Wallis test and Dunn post-test; P < 0.01 for muscle 6/7, one-way ANOVA, followed by a Tukey–Kramer post-test; P < 0.001 for muscle 12, Kruskal–Wallis test and Dunn post-test). There are no significant changes in type I bouton numbers when wild-type controls are compared with either of the two KH domain mutants. The allele designations denote the sole source of dFMR1 protein. Results are from analysis of at least 20 hemi-segments for each genotype.
Analysis of midline crossing frequency in mushroom-body β-lobe neurons:
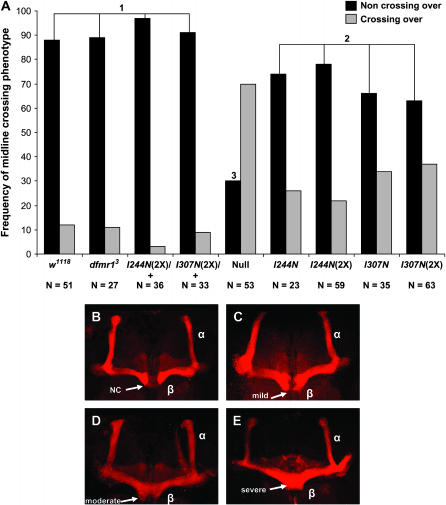
Axon development defects have been reported in the central nervous system of flies homozygous for null alleles of dfmr1. The ventral lateral neurons, dorsal cluster neurons, and neurons of the mushroom body all have visible defects in branching, neurite extension, and/or guidance (Dockendorff et al. 2002; Morales et al. 2002; Michel et al. 2004; Pan et al. 2004; Reeve et al. 2005). A significant fraction of flies homozygous for strong or null alleles of dfmr1 have midline crossings of MB β-lobe neurons (Michel et al. 2004). To monitor the effects of the KH domain alleles on axon development, we used the mushroom-body β-lobe phenotype described by Michel et al. (2004), visualizing MBs in whole-brain mounts from 2-day-old animals via anti-FasII immunostaining. Figure 3 shows that, when compared to a wild-type allele control, flies expressing dFMR1 with the I244N or I307N substitution had a significant increase in the frequency of midline crossings compared to controls where a wild-type allele of dfmr1 was present, but this frequency was not as great as that observed in brains from flies homozygous for a null allele of dfmr1 (Figure 3). Thus, the KH domains of dFMR1 play a role in regulating processes that contribute to normal axon development, and the other dFMR1 domains must contribute functions as well. We also examined the midline crossing phenotype in brains from flies where the dosage of either KH domain allele was increased. The frequency of midline crossings did not change significantly from what was observed with the single dose, indicating that the Ile → Asn substitutions have a strong effect on the function of the KH domains and that other domains of dFMR1 are not able to compensate for the defects.
Figure 3.—
MB β-lobe phenotypes of flies with dfmr1 KH domain alleles. (A) Representation of the frequency with which a midline crossing of β-lobe neurons was observed. Anti-FasII staining of MBs from 2-day-old flies shows that the I244N and I307N substitutions result in a frequency of midline crossing phenotypes intermediate to flies with a wild-type allele of dfmr1 and to those homozygous for a dfmr1 null allele. The genotypes denote the allele of dfmr1 being expressed, while the presence of a wild-type allele to test for dominant effects of the mutant allele is denoted by a “+.” The frequency with which a no-crossing phenotype occurred did not change upon an increase in dosage of either mutant KH domain protein. Expression of either mutant KH domain transgene in a background with a wild-type copy of dfmr1 present has no effect on midline crossing frequency, indicating that the transgene insertions and mutant proteins do not elicit a detectable dominant effect. Genotypes grouped under a common numerical designation do not differ from each other in percentage of brains observed with a midline crossing of β-lobe axons, while those under different numerical designations differ from each other as judged by chi-square tests of homogeneity. KH domain alleles differ from the null allele in frequency of midline crossing (P < 0.0001) and from flies with a wild-type allele of dfmr1 (P = 0.0152). (B–E) Representative examples of MB morphology illustrating the variety of midline crossing phenotypes observed. α- and β-Lobes are noted, while arrows point to the midline where crossovers of the β-lobe neurons may occur. NC, no crossover.
The KH domain mutations display a partially penetrant circadian phenotype in constant darkness:
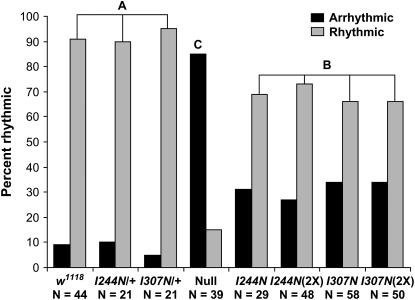
Flies with null alleles of dfmr1 fail to retain circadian rhythmicity when transferred into constant darkness (DD) with ∼80% penetrance (Dockendorff et al. 2002; Inoue et al. 2002; Morales et al. 2002). The KH domain mutants had circadian behavior examined by monitoring rest/activity rhythms in both LD and DD. Flies with either mutant allele are capable of responding to light, as judged by their rhythmic locomotion activity (T. Dockendorff and J. Park, unpublished observations). As was seen with the MB β-lobe phenotype, both KH domain substitutions resulted in a statistically significant increase in the percentage of flies that fail to retain rhythmic locomotion activity in constant darkness when compared to flies with a wild-type allele of dfmr1 (Figure 4). Likewise, the percentage of KH domain mutants lacking rhythmic activity is not as great as that observed for flies homozygous for the dfmr13 null allele, demonstrating that the KH domain alleles result in partial loss of function. For mutant flies determined to have retained rhythmic locomotion activity in constant darkness, the DD period of such flies did not significantly differ from flies with a wild-type dfmr1 allele (not shown). Flies expressing both the mutant transgene allele and a wild-type allele of dfmr1 do not differ from wild-type controls in the percentage of animals judged to have retained rhythmic locomotion, demonstrating that the insertions and mutant alleles have no dominant effect. An increase in dosage of either KH domain allele had no effect on the percentage of flies judged to be arrhythmic, again demonstrating a strong loss-of-function effect on the KH domains and the inability of other dFMR1 domains to provide compensatory function for this phenotype.
Figure 4.—
Analysis of circadian locomotion activity of flies expressing dFMR1 with mutant KH domains in constant darkness. An assignment of rhythmic vs. arrhythmic activity for individual flies was determined using ClockLab software as described in materials and methods. The percentage of flies from each genotype judged to be rhythmic was compared by a chi-square test for homogeneity. Genotypes that are grouped by a common number do not have any significant difference between them in the percentage of animals displaying a rhythmic locomotion phenotype, while separate groups differ to a confidence level of <0.0001. Increasing the dose of mutant dFMR1 protein had no significant effect on the percentage of animals judged to have maintained rhythmic locomotion activity, indicating that other RNA-binding domains of the mutant proteins are unable to compensate for the defects in the KH domains. The genotypes denote the allele of dfmr1 that is the sole source of dFMR1 protein, while the presence of a wild-type allele to test for dominant effects of the mutant allele is denoted by a “+.” Flies that express both a mutant and wild-type allele of dfmr1 resemble wild-type flies in the percentage of animals judged to be rhythmic, demonstrating that the mutant allele and transgene insertion do not have a detectable dominant effect.
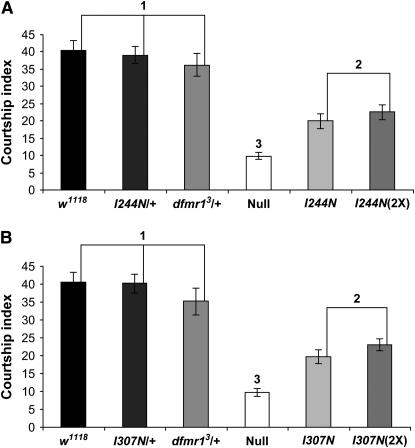
Naive courtship activity is reduced in flies with either KH domain mutation:
Courtship in Drosophila consists of stereotyped behaviors by males toward receptive females (reviewed by Hall 1994; Greenspan and Ferveur 2000). The courtship process has thus been used as an ethologically relevant measure of Drosophila behavior. Flies with a null allele of dfmr1 have deficits in naive courtship activity (Dockendorff et al. 2002), which is measured as the percentage of time that a male fly spends in courtship activity toward a receptive virgin female during a given period of time and defined as the courtship index. We analyzed the naive courtship activity of flies with the KH domain alleles, measuring the time spent by the male in following the female target, wing extension and vibration, tapping with foreleg, and attempted copulation (Figure 5). Flies that harbor both a copy of the transgene bearing either of the KH domain alleles and a wild-type allele of dfmr1 do not significantly differ from a wild-type control in the amount of time engaged in courtship behavior, showing that these insertions and alleles do not result in any dominant effect that contributes to the phenotype (Figure 5, A and B). Our analyses of courtship activity in flies where the KH domain allele is the only source of dFMR1 show that the Ile → Asn substitutions in either KH domain has a significant adverse effect on the courtship index when compared to flies with a wild-type allele. The values for the courtship index of both KH domain mutants are also significantly different from those seen with the null allele of dfmr1, indicating that dFMR1 proteins with the KH domain point mutations still have activity and behave as partial loss-of-function alleles. An increase in the dosage of either KH domain allele results in only a small, statistically insignificant increase in the courtship index. This increase could be accounted for by elevated expression of the mini white marker associated with the second copy of the P-element vector, since the white gene is known to positively influence courtship activity (reviewed by Hall 1994). Thus, as was seen with the morphology of mushroom bodies and circadian locomotion, an increased dosage of the mutant KH domain alleles did not provide rescue of this phenotype.
Figure 5.—
Analysis of naive courtship activity of flies expressing dFMR1 KH domain mutations. Naive courtship was analyzed as described in materials and methods. At least 25 flies of each genotype were tested. For each mutant KH domain transgene, expression in a background with a wild-type allele of dfmr1 does not result in a phenotype differing from wild type, indicating that the transgene insertion and mutant protein do not induce a detectable dominant effect. (A) Flies expressing dFMR1 with the I244N substitution as the sole source of dFMR1 protein have a significant decrease in naive courtship activity compared to flies with a wild-type allele of dfmr1, but the decrease in courtship activity is not as strong as is observed in flies homozygous for a dfmr1 null allele. Increasing the dosage of the I244N allele does not result in a significant increase in courtship activity. Courtship indexes were arcsin transformed and the data were analyzed by a one-way ANOVA, followed by a Tukey–Kramer post-test. The P-value for the ANOVA is <0.0001. Genotypes under the same numerical heading do not vary from each other to a significant extent, while genotypes under different numerical headings are significantly different from each other (P < 0.05). (B) The I307N substitution results in a significant decrease in naive courtship activity compared to flies expressing wild-type dFMR1, but not to the degree observed with flies homozygous for a null allele of dfmr1. As was seen with the I244N allele, an increase in dosage of the I307N allele does not produce a significant increase in naive courtship activity. The data for these genotypes were processed in the same manner as the I244N flies, and the P-value for the ANOVA is <0.0001. Genotypes under the same numerical grouping do not differ from each other in courtship activity to a significant extent, while genotypes under different numerical groupings have a significant variation in the courtship index (P < 0.01).
DISCUSSION
The vast majority of human fragile X cases arise through the expansion of a trinucleotide repeat, resulting in silencing of the FMR1 gene. Thus, relatively little insight into structure–function relationships has been gained from analysis of human FMR1 alleles. Biochemical and cell-culture-based studies of fragile X proteins have uncovered several RNA-binding domains that likely contribute to their in vivo function. The amenability of Drosophila melanogaster to transgenics provides an opportunity to conduct structure–function studies of FMRP in the context of an intact animal. To this end, we have generated flies expressing dfmr1 alleles where a codon for a highly conserved isoleucine residue in each of the two KH domains was mutated to code for asparagine. These mutations are predicted to result in a strong loss of affinity for specific RNA ligands as judged from structural (Lewis et al. 2000; Ramos et al. 2003) and biochemical studies (Darnell et al. 2005) and may interfere with the ability of FMRP to interact with other proteins as well (Feng et al. 1997; Laggerbauer et al. 2001; Ramos et al. 2003). We then examined the effects of these mutations on neural development and behavior phenotypes that are associated with null alleles of dfmr1. Several conclusions can be made from these results. Neither of the KH domain alleles produced a phenotype that matched the degree of severity seen with the dfmr1 deletion null allele. For all phenotypes analyzed, the KH domain alleles were recessive to the wild-type dfmr1 allele. The failure of increased dosage of the mutant proteins to provide any significant measure of rescue indicates that the Ile → Asn substitutions are strong loss-of-function mutations in the KH domains, which is consistent with past biochemical and biophysical analyses of KH domains (Lewis et al. 2000; Ramos et al. 2003; Darnell et al. 2005). These results suggest that the ability of the mutant proteins to bind certain RNA species in vivo is lost and that other RNA-binding domains of dFMR1 cannot compensate for the defect. Prior studies have shown that the individual FMRP RNA-binding domains can bind RNA as a discrete unit in vitro (Adinolfi et al. 1999; Darnell et al. 2001, 2005; Schaeffer et al. 2001), and our results are consistent with the observations from these studies. The partial loss-of-function phenotypes resulting from these KH domain alleles demonstrate that the mutant proteins retain function and must be able to bind RNA and assemble into at least some mRNP complexes in vivo.
KH domain phenotypes and roles for other RNA-binding domains of FMRPs:
Why is there a difference between the lack of phenotype seen with the larval NMJ bouton numbers and the partial loss of function observed for other phenotypes analyzed? It is possible that regulation of a different subset of RNAs is involved in larval NMJ bouton development and that these RNAs are not reliant upon interaction with dFMR1 KH domains to conduct their functions and be appropriately regulated. A number of possibilities can explain the partial loss-of-function phenotypes associated with mushroom-body development, courtship behavior, and circadian locomotion activity. It could be that dFMR1 regulates the activity of multiple genes involved in these processes and that the KH domains are responsible for modulating the activity of a subset of these genes. Multiple RNA-binding domains in dFMR1 could also regulate the activity of any one transcript, and loss of KH domain function could lead to a partial degree of misregulation for such transcripts that result in the degree of phenotype observed.
That we did not observe a null phenotype with either of the KH domain alleles makes it rather probable that the other RNA-binding domains of FMRP significantly contribute to its in vivo function. Along with the G-quartet binding RGG box, the N-terminal 217 amino acids of human FMRP also binds RNA (Adinolfi et al. 1999, 2003; Zalfa et al. 2005). The N-terminal amino acids of FMRP are well conserved between mammals and insects (Wan et al. 2000) and are related to methyl-substrate-binding domains that are associated with chromatin regulation (Maurer-Stroh et al. 2003). This is of interest because FMRP can be detected in the nucleus and has biochemical and genetic interactions with Argonaute proteins (Verheij et al. 1993; Caudy et al. 2002; Ishizuka et al. 2002; Jin et al. 2004b; Xu et al. 2004). Mutations of RNA interference (RNAi) components affect silencing of heterochromatin in D. melanogaster (Pal-Bhadra et al. 2004; see Lippman and Martienssen 2004; Matzke and Birchler 2005; Wassenegger 2005 for reviews of RNAi-based regulation of chromatin). Indeed, it has recently been reported that a null allele of dfmr1 affects white gene expression in centromeric heterochromatin (Deshpande et al. 2006). If an FMRP is part of a complex that modulates chromatin structure via RNAi-based mechanisms, the loss of such activity via a null mutation could conceivably affect the expression of many genes that might be part of the fragile X pathway, including those that may contribute to the phenotypes examined in this study.
Comparing phenotypes of the human I304N and Drosophila I307N substitutions:
The failure of the I304N protein to bind certain RNAs and its abnormal mRNP fractionation profile have been suggested as the basis for the unusually severe fragile X phenotypes observed with a patient expressing the I304N substitution (Feng et al. 1997; Darnell et al. 2005) and have been hypothesized to arise from a dominant effect exerted by the I304N protein (Feng et al. 1997). The studies here provide an opportunity to make comparisons between the human I304N and Drosophila I307N phenotypes. Given the severity of fragile X phenotypes associated with the I304N substitution, our findings of relatively modest phenotypes associated with the analogous I307N substitution in the Drosophila model may seem surprising. The failure to observe a phenotype with the I307N protein that matches the strength of a dfmr1 null allele means that the I307N protein must be able to bind RNA and to assemble into at least a subset of the mRNP complexes that the wild-type protein does. We feel that several hypotheses are plausible for explaining the differences observed between the human I304N and the Drosophila I307N phenotypes. Subtle differences in substrate binding by these KH domains is one possibility, with the human I304N protein unable to bind certain critical RNA substrates, while the binding of orthologous substrates (if they exist) to the Drosophila I307N protein is impaired to a lesser degree. In vertebrates, a FMRP interacts with FXR1 and FXR2 proteins that are not present in D. melanogaster (Zhang et al. 1995). Since the I304N protein can interact with both FXR1 and FXR2 in vitro (Laggerbauer et al. 2001; Mazroui et al. 2003), it is possible that I304N FMRP could induce a deleterious gain-of-function effect on either FXR protein that enhances the severity of the phenotype. Alternatively, since not all I304N protein cofractionates with FXR2 (Feng et al. 1997), the I304N protein may acquire a deleterious function when not in complex with FXR2. Another consideration is the possibility of unusual contributions from the genetic background of the I304N individual that could enhance the fragile X phenotypes. To recapitulate the I304N substitution in the mouse model could be helpful in discerning whether these final two hypotheses have validity. Finally, none of the above explanations are mutually exclusive, and thus any combination of these scenarios might contribute to the severity of the fragile X phenotypes observed with the I304N patient.
The KH domain alleles as probes for FMRP functions:
What RNAs do fragile X proteins bind to? What is the composition of a fragile X protein-containing mRNP particle? These are questions that still dominate research on FMRP. Biochemical screens utilizing microarrays, along with yeast two-hybrid screens and proteomics approaches, have been undertaken to address these questions (Ceman et al. 1999; Brown et al. 2001; Darnell et al. 2001; Schenck et al. 2001; Ishizuka et al. 2002; Miyashiro et al. 2003; Costa et al. 2005; Zarnescu et al. 2005) and have uncovered several interacting proteins and dozens of RNA species identified as candidate ligands. In vivo genetics-based analyses of these interactions will be necessary for their final validation. Drosophila has proven to be a significant model for study of fragile X protein function owing to the similarities with vertebrate models in biochemical properties and loss-of-function phenotypes (Wan et al. 2000; Zhang et al. 2001; Dockendorff et al. 2002; McBride et al. 2005). Thus, the tools of Drosophila genetics will be instrumental in identifying and ordering the physiologic pathways that fragile X protein regulates. In addition to their value as stocks with sensitized backgrounds for probing genetic interactions, these stocks with the KH domain mutations will serve as useful comparative tools for biochemical and genetic studies in identifying the RNAs regulated by the fragile X protein and the protein interactions needed for its role in neural function.
Acknowledgments
We thank Richard Edelman of the Miami University Electron Microscopy Facility for training and technical expertise with microscopy. Michael Bradley and Courtney Doughty assisted with courtship behavior studies, and Sean McBride and Kathy Siwicki provided useful tips for conducting these assays. Thanks go to Nancy Solomon and Michael Hughes for discussions and assistance with statistical analyses. The University of Iowa Developmental Studies Hybridoma Bank provided antibodies used in this study. P.B. is the recipient of a grant-in-aid to promote research from Sigma Xi, and C.F.F. and M.C.J. were supported by Summer Undergraduate Research Fellowships from the Howard Hughes Medical Institute and Miami University. This research was supported by National Institutes of Health grants MH66197 to J.H.P., MH067622-01 to J.J.F., and GM068468 to T.C.D.
References
- Adinolfi, S., C. Bagni, G. Musco, T. Gibson, L. Mazzarella et al., 1999. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 5: 1248–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adinolfi, S., A. Ramos, S. R. Martin, F. Dal Piaz, P. Pucci et al., 2003. The N-terminus of the fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry 42: 10437–10444. [DOI] [PubMed] [Google Scholar]
- Ashley, C. T., Jr., K. D. Wilkinson, D. Reines and S. T. Warren, 1993. FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262: 563–566. [DOI] [PubMed] [Google Scholar]
- Bagni, C., and W. T. Greenough, 2005. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 6: 376–387. [DOI] [PubMed] [Google Scholar]
- Bardoni, B., and J. L. Mandel, 2002. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev. 12: 284–293. [DOI] [PubMed] [Google Scholar]
- Brown, V., P. Jin, S. Ceman, J. C. Darnell, W. T. O'Donnell et al., 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487. [DOI] [PubMed] [Google Scholar]
- Caudy, A. A., M. Myers, G. J. Hannon and S. M. Hammond, 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman, S., V. Brown and S. T. Warren, 1999. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol. Cell. Biol. 19: 7925–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, A., Y. Wang, T. C. Dockendorff, H. Erdjument-Bromage, P. Tempst et al., 2005. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev. Cell 8: 331–342. [DOI] [PubMed] [Google Scholar]
- Darnell, J. C., K. B. Jensen, P. Jin, V. Brown, S. T. Warren et al., 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107: 489–499. [DOI] [PubMed] [Google Scholar]
- Darnell, J. C., C. E. Fraser, O. Mostovetsky, G. Stefani, T. A. Jones et al., 2005. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 19: 903–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle, K., A. J. Verkerk, E. Reyniers, L. Vits, J. Hendrickx et al., 1993. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 3: 31–35. [DOI] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun and P. Schedl, 2006. The Drosophila fragile X protein dFMR1 is required during early embryogenesis for pole cell formation and rapid nuclear division cycles. Genetics 174: 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff, T. C., H. S. Su, S. M. J. McBride, Z. Yang, C. H. Choi et al., 2002. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 34: 973–984. [DOI] [PubMed] [Google Scholar]
- Dölen, G., and M. F. Bear, 2005. Courting a cure for fragile X. Neuron 45: 642–644. [DOI] [PubMed] [Google Scholar]
- Feng, Y., D. Absher, D. E. Eberhart, V. Brown, H. E. Malter et al., 1997. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1: 109–118. [DOI] [PubMed] [Google Scholar]
- Gao, F.-B., 2002. Understanding fragile X syndrome: insights from retarded flies. Neuron 34: 859–862. [DOI] [PubMed] [Google Scholar]
- Greenspan, R. J., and J. F. Ferveur, 2000. Courtship in Drosophila. Annu. Rev. Genet. 34: 205–232. [DOI] [PubMed] [Google Scholar]
- Hall, J. C., 1994. The mating of a fly. Science 264: 1702–1714. [DOI] [PubMed] [Google Scholar]
- Inoue, S., M. Shimoda, I. Nishinokubi, M. C. Siomi, M. Okamura et al., 2002. A role for the Drosophila fragile X-related gene in circadian output. Curr. Biol. 12: 1331–1335. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., M. C. Siomi and H. Siomi, 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P., and S. T. Warren, 2003. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem. Sci. 28: 152–158. [DOI] [PubMed] [Google Scholar]
- Jin, P., R. S. Alisch and S. T. Warren, 2004. a RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 6: 1048–1053. [DOI] [PubMed] [Google Scholar]
- Jin, P., D. C. Zarnescu, S. Ceman, M. Nakamoto, J. Mowrey et al., 2004. b Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7: 113–117. [DOI] [PubMed] [Google Scholar]
- Laggerbauer, B., D. Ostareck, E. M. Keidel, A. Ostareck-Lederer and U. Fischer, 2001. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10: 329–338. [DOI] [PubMed] [Google Scholar]
- Lee, A., W. Li, K. Xu, B. A. Bogert, K. Su et al., 2003. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130: 5543–5552. [DOI] [PubMed] [Google Scholar]
- Lewis, H. A., K. Musunuru, K. B. Jensen, C. Edo, H. Chen et al., 2000. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., and R. Martienssen, 2004. The role of RNA interference in heterochromatic silencing. Nature 431: 364–370. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., and J. A. Birchler, 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6: 24–35. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh, S., N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber et al., 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28: 69–74. [DOI] [PubMed] [Google Scholar]
- Mazroui, R., M. E. Huot, S. Tremblay, N. Boilard, Y. Labelle, et al., 2003. Fragile X mental retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 12: 3087–3096. [DOI] [PubMed] [Google Scholar]
- McBride, S. M. J., C. H. Choi, Y. Wang, D. Liebelt, E. Braunstein et al., 2005. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron 45: 753–764. [DOI] [PubMed] [Google Scholar]
- Michel, C. I., R. Kraft and L. L. Restifo, 2004. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J. Neurosci. 24: 5798–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro, K. Y., A. Beckel-Mitchener, T. P. Purk, K. G. Becker, T. Barret et al., 2003. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37: 417–431. [DOI] [PubMed] [Google Scholar]
- Morales, J., P. R. Hiesinger, A. J. Schroeder, K. Kume, P. Verstreken et al., 2002. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron 34: 961–972. [DOI] [PubMed] [Google Scholar]
- Musco, G., G. Stier, C. Joseph, M. A. C. Morelli, M. Nilges et al., 1996. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell 85: 237–245. [DOI] [PubMed] [Google Scholar]
- Musco, G., A. Kharrat, G. Stier, F. Fraternali, T. J. Gibson et al., 1997. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 4: 712–716. [DOI] [PubMed] [Google Scholar]
- O'Donnell, W. T., and S. T. Warren, 2002. A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25: 315–338. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Pan, L., Y. Q. Zhang, E. Woodruff and K. Broadie, 2004. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr. Biol. 14: 1863–1870. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V., 1988. Vectors for P-mediated transformation in Drosophila. Biotechnology 10: 437–456. [DOI] [PubMed] [Google Scholar]
- Ramos, A., D. Hollingworth and A. Pastore, 2003. The role of a clinically important mutation in the fold and RNA-binding properties of KH motifs. RNA 9: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve, S. P., L. Bassetto, G. K. Genova, Y. Kleyner, M. Leyssen et al., 2005. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr. Biol. 15: 1156–1163. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaeffer, C., B. Bardoni, J. L. Mandel, B. Ehresmann, C. Ehresmann et al., 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 20: 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck, A., B. Bardoni, A. Moro, C. Bagni and J. L. Mandel, 2001. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. USA 98: 8844–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., M. C. Siomi, R. L. Nussbaum and G. Dreyfuss, 1993. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74: 291–298. [DOI] [PubMed] [Google Scholar]
- Siomi, H., M. Choi, M. C. Siomi, R. L. Nussbaum and G. Dreyfuss, 1994. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77: 33–39. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Verheij, C., C. E. Bakker, E. de Graaff, J. Keulemans, R. Willemsen et al., 1993. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature 363: 722–724. [DOI] [PubMed] [Google Scholar]
- Wan, L., T. C. Dockendorff, T. A. Jongens and G. Dreyfuss, 2000. Characterization of dFMR1, a Drosophila homolog of the fragile X mental retardation protein. Mol. Cell. Biol. 20: 8536–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., A. Iacoangeli, D. Lin, K. Williams, R. B. Denman et al., 2005. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 171: 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., 2005. The role of the RNAi machinery in heterochromatin formation. Cell 122: 13–16. [DOI] [PubMed] [Google Scholar]
- Xu, K., B. A. Bogert, W. Li, K. Su, A. Lee et al., 2004. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein pickpocket1. Curr. Biol. 14: 1025–1034. [DOI] [PubMed] [Google Scholar]
- Zalfa, F., S. Adinolfi, I. Napoli, E. Kuhn-Holsken, H. Urlaub et al., 2005. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J. Biol. Chem. 280: 33403–33410. [DOI] [PubMed] [Google Scholar]
- Zarnescu, D. C., P. Jin, J. Betschinger, M. Nakamoto, Y. Wang et al., 2005. Fragile X protein functions with lgl and the par complex in flies and mice. Dev. Cell 8: 43–52. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. Q., and K. Broadie, 2005. Fathoming fragile X in fruit flies. Trends Genet. 21: 37–45. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., J. P. O'Connor, M. C. Siomi, S. Srinivasan, A. Dutra et al., 1995. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14: 5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. Q., A. M. Bailey, H. J. G. Matthies, R. B. Renden, M. A. Smith et al., 2001. Drosophila fragile X-related gene regulates the MAP1B homolog futsch to control synaptic structure and function. Cell 107: 591–603. [DOI] [PubMed] [Google Scholar]