Abstract
The TOR protein kinases exhibit a conserved role in regulating cellular growth and proliferation. In the fission yeast two TOR homologs are present. tor1+ is required for starvation and stress responses, while tor2+ is essential. We report here that Tor2 depleted cells show a phenotype very similar to that of wild-type cells starved for nitrogen, including arrest at the G1 phase of the cell cycle, induction of nitrogen-starvation-specific genes, and entrance into the sexual development pathway. The phenotype of tor2 mutants is in a striking contrast to the failure of tor1 mutants to initiate sexual development or arrest in G1 under nitrogen starvation conditions. Tsc1 and Tsc2, the genes mutated in the human tuberous sclerosis complex syndrome, negatively regulate the mammalian TOR via inactivation of the GTPase Rheb. We analyzed the genetic relationship between the two TOR genes and the Schizosaccharomyces pombe orthologs of TSC1, TSC2, and Rheb. Our data suggest that like in higher eukaryotes, the Tsc1–2 complex negatively regulates Tor2. In contrast, the Tsc1–2 complex and Tor1 appear to work in parallel, both positively regulating amino acid uptake through the control of expression of amino acid permeases. Additionally, either Tsc1/2 or Tor1 are required for growth on a poor nitrogen source such as proline. Mutants lacking Tsc1 or Tsc2 are highly sensitive to rapamycin under poor nitrogen conditions, suggesting that the function of Tor1 under such conditions is sensitive to rapamycin. We discuss the complex genetic interactions between tor1+, tor2+, and tsc1/2+ and the implications for rapamycin sensitivity in tsc1 or tsc2 mutants.
TOR proteins, the central protein kinases that control cell growth and proliferation, were first identified in the yeast Saccharomyces cerevisiae as the targets for the immunosuppressive and potential anticancerous drug rapamycin (Wullschleger et al. 2006). Rapamycin binds the small FKBP12 protein and the resulting protein–drug complex binds and inhibits TOR-dependent activities. Two TOR homologs are present in S. cerevisiae, TOR1 and TOR2. These are found at the core of two different evolutionary conserved complexes (Loewith et al. 2002). Although a single TOR gene is found in higher eukaryotes, two distinct physical and functional TOR complexes are also present in mammalian and Drosophila cells (Kim et al. 2003; Jacinto et al. 2004; Sarbassov et al. 2004). The mammalian mTORC1 complex regulates cell growth, in part, via phosphorylation of the two translation initiation factors, p70 S6 kinase and 4E-BP1 proteins (Hay and Sonenberg 2004; Tee and Blenis 2005). mTORC2 regulates the actin cytoskeleton (Jacinto et al. 2004) and the cell growth and survival regulator Akt/PKB (Sarbassov et al. 2005).
In accord with the suggestion that TOR regulates cell growth in response to nutrient availability, S. cerevisiae cells lacking both TOR1 and TOR2 exhibit many features characteristics of starved cells (Barbet et al. 1996; Beck and Hall 1999; Di Como and Arndt 1996; Noda and Ohsumi 1998; Zaragoza et al. 1998). Similarly, loss of function of the TOR proteins in worms, flies, and mice results in phenotypes like those of cells suffering from nutrient deprivation (Oldham et al. 2000; Zhang et al. 2000; Long et al. 2002; Gangloff et al. 2004; Murakami et al. 2004).
In higher eukaryotes, TOR proteins are negatively regulated by the TSC1–TSC2 heterodimer. Mutations in either TSC1 or TSC2 cause a human syndrome, known as tuberous sclerosis complex (TSC), which is characterized by benign tumors and severe neurological defects. TSC2 encodes a GTPase-activating protein (GAP) and together with TSC1 converts the small GTPase Rheb into its GDP-bound inactive form. Rheb binds mTOR and positively regulates its activity (Manning and Cantley 2003; Li et al. 2004). More recent results demonstrated that TSC1/2 also has positive effects on TOR signaling, suggesting the presence of a negative feedback loop (Harrington et al. 2004; Shah et al. 2004; Um et al. 2004).
The fission yeast, but not the budding yeast, contains homologs for TSC1/2, known as tsc1+ and tsc2+. Schizosaccharomyces pombe Tsc1 and Tsc2 proteins also work as a complex (Matsumoto et al. 2002) and missense mutations that cause TSC in humans abolish the activity of the S. pombe homologs (Van Slegtenhorst et al. 2004). Disruption of tsc1+ or tsc2+ leads to amino acid uptake defects, impairs sexual development, and affects gene induction upon nitrogen starvation (Matsumoto et al. 2002; Van Slegtenhorst et al. 2004; Nakase et al. 2006). Several findings demonstrate that the Tsc1–2 complex negatively regulates Rhb1, the S. pombe Rheb homolog: rhb1 mutants exhibit a phenotype opposite to that of tsc1/2 (Yang et al. 2001; Urano et al. 2005) and mutations that impair Rhb1 activity can reverse the amino acid uptake defect in tsc1/2 mutants (Van Slegtenhorst et al. 2004; Nakase et al. 2006). The rhb1+ gene is essential and cells disrupted for rhb1+ arrest at the G1 phase of the cell cycle with the morphology and physiology of nitrogen-starved cells (Mach et al. 2000; Yang et al. 2001). The nitrogen-starved phenotype of rhb1 mutants suggests that rhb1+ participates in signaling nitrogen availability.
The S. pombe tor1+ gene is required under starvation and stress conditions (Kawai et al. 2001; Weisman and Choder 2001). Δtor1 cells die quickly once they exit the logarithmic phase and do not initiate sexual development in response to starvation conditions. Δtor1 cells are also highly sensitive to osmotic and oxidative stress. Given the conserved role of TOR proteins in regulating growth, the finding that Tor1 is required mainly under starvation, when little growth is required, is surprising. However, Tor1 exerts its functions in stress and starvation via phosphorylation of the S. pombe p70 S6 kinase homolog, gad8+ (Matsuo et al. 2003). Thus, the substrate and mode of action of Tor1 is highly conserved in evolution. Rapamycin does not inhibit growth, entrance into stationary phase, or stress responses in fission yeast (Weisman et al. 1997; Weisman 2004). The only rapamycin-sensitive TOR-dependent function reported to date in S. pombe is amino acid uptake, which is mediated via a Tor1-dependent function (Weisman et al. 2005).
In contrast to our detailed knowledge on tor1+ activity, no description of Tor2 loss of function has so far been reported. Here we show that disruption of tor2+ results in a phenotype that highly resembles that of nitrogen-starved cells and that the Tsc1/2–Rhb1 module has different effects on Tor1 and Tor2 functions. While loss of function of Tor2 highly resembles that of rhb1 mutants, loss of function of Tsc1/2 shares similar defects with Tor1 disruption. We discuss the complex genetic interactions between tor1+, tor2+, and tsc1/2+.
MATERIALS AND METHODS
Yeast strains, media, and general techniques:
Yeast strains are described in Table 1. Growth media were prepared as in Moreno et al. (1991). YES is YE supplemented with 75 μg/ml adenine and uracil. EMM (Edinburgh minimal medium) is defined as medium in which the sole nitrogen source is ammonium chloride (Stettler et al. 1996). Minimal medium was supplemented as required. Leucine, histidine, adenine, and uracil were supplemented at 75 μg/ml, unless otherwise indicated. For proline (Pro) and glutamic acid (Glu) plates, ammonium chloride in the EMM medium was replaced with 10–30 mm proline and 20 mm glutamic acid, respectively (Young and Fantes 1987). EMM − N contains no nitrogen. Rapamycin was used at a final concentration of 100 ng/ml as described previously (Weisman et al. 1997). Transformation of S. pombe cells was performed by electroporation (Prentice 1992). Assays for mating efficiency were carried out as described (Weisman et al. 1997).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| TA001 | 972 h− | Lab stock |
| TA007 | leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 h−/h+ | Lab stock |
| TA016 | ade6-M216 leu1-32 ura4-D18 h90 | Lab stock |
| TA157 | tor1∷ura4+ade6-M216 leu1-32 ura4-D18 h90 | Weisman and Choder (2001) |
| TA313 | tor2∷ura+ura4-D18 leu1-32 ade6 pREP81-tor2+h− | This study |
| TA390 | tor1∷ura4+ | Weisman and Choder (2001) |
| TA417 | leu1-32 ura4-D18 ade6-216 his7-366 h− | M. van Slegtenhorst |
| TA418 | tsc2∷kan+leu1-32 ura4-D18 ade6-216 his7-366 h− | M. van Slegtenhorst |
| TA419 | tsc1∷kan+leu1-32 ura4-D18 ade6-210 his7-366 h+ | M. van Slegtenhorst |
| TA426 | tor1∷ura4+ tor2∷his1+his1-102 ura4-D18 leu1-32 ade6-M210 pREP81-tor2+h− | This study |
| TA430 | tor2∷ura+ura4-D18 leu1-32 ade6 pREP81-tor2+h90 | This study |
| TA434 | rhb1+/rhb∷ura4+ade6-M210/ade-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 | This study |
| TA443 | rhb1∷ura4+ade6 ura4-D18 leu1-32 pREP81-rhb1+ | This study |
| TA449 | tsc1∷G418 | This study |
| TA450 | tsc2∷G418 | This study |
| TA459 | tor1∷ura4+tsc2∷kan+ura4-D18 | This study |
| TA467 | tsc1∷G418 fkh1∷ura4+ | This study |
| TA468 | tsc2∷G418 fkh1∷ura4+ | This study |
| TA481 | tor1∷ura4+tsc1∷kan+ura4-D18 | This study |
| TA502 | tor2∷his1+his1-102 ura4-D18 leu1-32 ade6-M210 pREP1-tor2+h90 | This study |
| TA561 | tor1∷ura4+ade6 leu1-32 ura4D-18 h− pREP81-tor2+ | This study |
| TA568 | leu1-32 pREP1-tor2+h− | This study |
| TA569 | tor1∷ura4+ tor2∷his1+his1-102 ura4-D18 leu1-32 ade6-M210 pREP1-tor2+h90 | This study |
| TA597 | fkh1∷ura4+ | This study |
tor2+ plasmid constructs:
For expression of tor2+ from its own promoter on a multicopy plasmid, a fragment of 7936 bp was amplified, using the Expand long template PCR system (Roche Applied Science). This fragment, containing the tor2+ ORF flanked by 413 and 509 bp, was cloned into the LEU2-based plasmid pIRT2 (Booher and Beach 1986). For expression under the regulation of the nmt1 promoter, a fragment of 7523 bp containing the tor2+ ORF and 509 bp downstream of it was amplified. This fragment was cloned into LEU1-based pREP1 or pREP81 plasmids (Maundrell 1993).
Measurement of leucine uptake:
Leucine uptake was performed as previously described in Weisman et al. (2005). Cells were grown to log phase in minimal medium. One-half milliliter of logarithmic cells was harvested and resuspended in 0.5 ml minimal medium containing 0.01 mm leucine together with 3H-labeled leucine (1–5 μCi of [l-4,5-3H(N)]-leucine, 50 Ci/mmol; New England Nuclear Life Science Products, Boston). Cells were incubated at 30° and samples were taken at 6 min and mixed with chilled minimal medium containing 10 mm leucine. Cells were then washed three times before being resuspended in water containing 0.5% SDS. Experiments were done in duplicates.
Northern blot analysis and semiquantitative RT–PCR:
Yeast RNA was extracted from logarithmic growing cells and the isp5+ and adh1+ mRNA were detected as previously described in Weisman et al. (2005). For semiquantitative RT–PCR analysis 50 ng yeast RNA were treated with DNAse I (Promega, Madison, WI) and used as template in one-step RT–PCR (QIAGEN, Valencia, CA). Samples were taken following 20, 25, and 30 cycles of 45 sec at 94°, 45 sec at 53°, and 90 sec at 72°.
Fluorescence-activated cell sorter analysis:
Cells were stained with propidium iodide and analyzed by a Becton Dickinson FACSort as described in Snaith and Forsburg (1999). Data were analyzed by Cell Quest software for Macintosh.
Disruption of S. pombe rhb1+:
A fragment containing 771 bp of the rhb1+ ORF and an additional 118 bp upstream of the stop codon was amplified by PCR from genomic DNA with primers P280 (GGAATTCCATATGGCTCCTATTAAATCTCGTAG) and P281 (TTTGTGTGCAAAGTATCACCTC) and subcloned into a pGEM-T vector (Promega) to give pGEMrhb1. A 350-bp ClaI–XhoI fragment of pGEMrhb1 was replaced with the ura4+ gene, resulting in the plasmid pGEMrhb1∷ura4+. Primers P280 and P281 were then used to amplify a 2.2-kb disruption fragment that was transformed into diploid TA007. Disruption was confirmed by PCR and Southern blot analysis.
RESULTS
Cells depleted for Tor2 arrest with a phenotype that highly resembles nitrogen-starved cells:
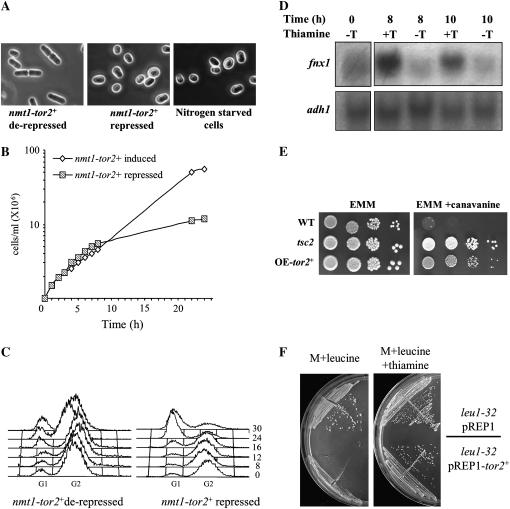
tor2+ is an essential gene (Weisman and Choder 2001). To further study the cellular role of tor2+, we cloned tor2+ under the regulation of the nmt1 promoter. This promoter is turned off in the presence of thiamine (Maundrell 1993). Cells carrying disruption of tor2+ at the chromosomal locus and expressing the nmt1-tor2+ construct were grown to midlog phase and then shifted to repressing conditions and samples were taken for cell number counting and cell morphology examination. As seen in Figure 1, A and B, cells shifted to the repressed conditions arrested after three to four divisions as small, rounded cells, similar to wild-type cells starved for nitrogen. Under nitrogen starvation, cells undergo mitosis at a shorter cell length and thus give rise to small cells (Fantes and Nurse 1977; Young and Fantes 1987). Nitrogen starvation of wild-type cells is also characterized by accumulation of cells in the G1 phase of the cell cycle (Fantes and Nurse 1977; Young and Fantes 1987). FACS analysis revealed that depletion of Tor2 leads to growth arrest with a G1 DNA content (Figure 1C), very similar to the arrest seen in wild-type cells following nitrogen starvation (Weisman and Choder 2001).
Figure 1.—
tor2 phenotypes. (A) Left and middle: tor2− cells expressing nmt1-tor2+ (TA313) were grown to midlog phase and transferred to fresh minimal medium in the absence (derepressed) or presence (repressed) of thiamine. After 12 hr, cells were visualized by light microscopy. Right: wild-type cells (TA001) were starved in minimal medium containing no nitrogen for 21 hr. (B) Δtor2 mutant cells (TA313) were grown as described above. At time zero thiamine was added and cells were counted. (C) FACS analysis of the cells sampled in B. (D) Northern blot analysis. Total RNA was prepared from samples taken at the indicated time points (hours). Northern blots were probed with fnx1+ and with adh1+ as a loading control. (E) Wild-type (TA001), Δtsc2 (TA450), and Δtor2 carrying nmt1-tor2+ (TA313) cells were grown in EMM to midlog phase. Four different dilutions were spotted on EMM plates with and without 60 μg/ml canavanine. Plates were incubated at 30° for 3 days. (F) Cells auxotrophic for leucine were transformed with vector only or with vector containing the nmt1-tor2+ construct. Slow-growth phenotype is observed only when the nmt1 promoter is derepressed (no thiamine). Cells were streaked on minimal (M) plates supplemented with 75 μg/ml leucine and incubated at 30° for 4 days.
We also found that the nitrogen-starved-like phenotype of Tor2-depleted cells and the arrest in G1 are very similar to the phenotype of cells depleted of Rhb1 (Mach et al. 2000). Cells depleted of Rhb1 are characterized by small cell size morphology, G1 arrest, and induction of the nitrogen-starvation-specific gene, fnx1+ (Mach et al. 2000). Northern blot analysis of total RNA from cells that were repressed for tor2+ revealed that fnx1+ mRNA was also increased when tor2+ was repressed (Figure 1D). Peak levels were seen 8 hr after tor2+ was repressed, at the time just before cell growth was dramatically reduced.
Another hallmark of the response to nitrogen starvation is initiation of sexual development, which is completely repressed in rich medium (Weisman 2004). Cells disrupted for tor2+ were patched on rich YE medium, which contains sufficient thiamine to repress transcription from the nmt1 promoter. Under these conditions no colonies are formed, but microscopic examination revealed that ∼30% of the cells underwent mating, meiosis, and sporulation. No meiosis was observed in isogenic wild-type cells, as expected. The ability of cells to enter sexual development on rich medium characterizes mutants of the cAMP pathway, such as cells lacking the adenylate cyclase, Cyr1/Git2, or the protein kinase A, Pka1 (Stiefel et al. 2004; Hoffman 2005). While the cAMP-dependent pathway is thought to regulate growth and developmental decisions mainly in response to glucose sensing, the nitrogen-starved phenotype of Tor2 depletion suggests that tor2+ regulates growth and development in response to nitrogen availability.
Overexpression of Tor2, like hyperactivation of Rhb1 or loss of function of Tsc1/2, results in resistance to canavanine:
Loss of function of Rhb1 results in a phenotype that resembles nitrogen starvation, while hyperactive Rhb1 mutants confer resistance to the arginine toxic analog canavanine, apparently because of downregulated uptake (Urano et al. 2005). We tested whether increasing Tor2 activity may also result in canavanine resistance. As shown in Figure 1E, overexpression of tor2+ induces canavanine resistance in wild-type cells. Thus, while rhb1 or tor2 mutants exhibit a nitrogen-starvation phenotype, their overactivation leads to canavanine resistance, consistent with the hypothesis that their overexpression results in signaling of nutrient/nitrogen sufficiency, leading to reduced uptake of nitrogenous compounds such as amino acids. Overexpression of tor2+ in cells that are auxotrophic for leucine leads to slow growth on minimal plates that are normally supplemented for leucine (Figure 1F). This phenotype is similar to loss of function of tsc1+ or tsc2+ (Matsumoto et al. 2002). The similarity between tor2 and rhb1 mutants and the opposite effects of tor2 and tsc1/2 mutants on leucine auxotrophs suggest that they work on the same signaling pathway and that like in higher eukaryotes rhb1+ positively regulate tor2+. This possibility is schematically presented in Figure 5C and is further discussed below.
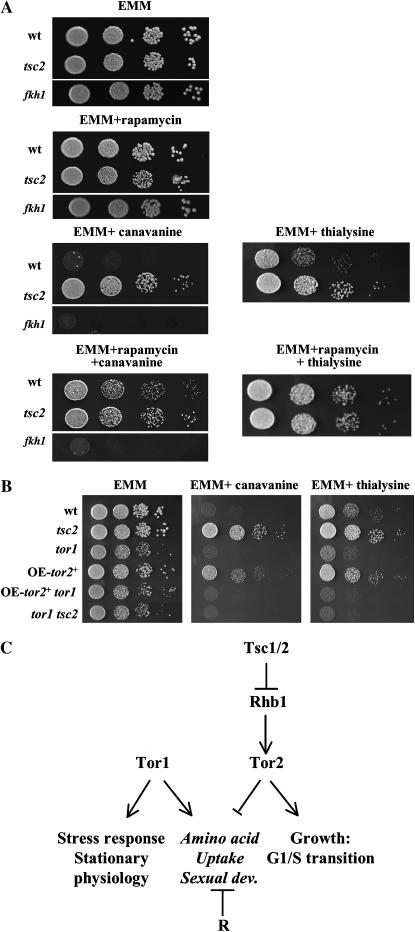
Figure 5.—
Rapamycin confers canavanine and thialysine resistance, while deletion of tor1+ confers hypersensitivity to these toxic amino acid analogs. (A) Serial dilutions of exponentially growing wild-type (TA001), Δtsc2 (TA450), or Δfkh1 (TA597) cells were spotted on EMM medium containing canavanine or thialysine in the absence or presence of rapamycin. (B) Serial dilutions of TA001, TA450, TA390, TA568, TA561, and TA459 were spotted on EMM plates in the presence of either canavanine or thialysine. (C) Working model. Tor2 positively regulates growth, allowing the G1/S transition under conditions of sufficient nutrients. Tor2 lies downstream of Rhb1 and is negatively regulated by Tsc1–2, while Tor1 acts in parallel with the Tsc1/2–Rhb1 module. The functions that are inhibited by rapamycin (R) are indicated in italics.
tor1 and tor2 null mutants exhibit opposite phenotypes with respect to response to nitrogen availability:
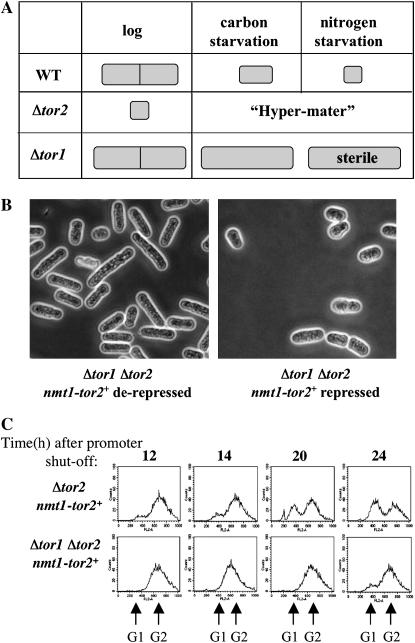
Under nitrogen starvation, tor1 mutants show an abnormal phenotype as cells remain elongated and do not accelerate entrance into mitosis, do not arrest in G1, and are completely sterile (Weisman and Choder 2001). In contrast, cells depleted of Tor2 are small, arrested in G1, and readily enter sexual development in the presence of rich medium. Thus, while Tor2-depleted cells appear as “always starved for nitrogen,” cells disrupted for tor1+ appear as “never starved for nitrogen” (Figure 2A). The opposite effects of two closely related homologs are clearly unexpected. An intriguing possibility is that the two homologs are connected to each other through a negative feedback loop. In such a scenario, absence of Tor1 may lead to overactivation of Tor2 and thus results in a phenotype of never starved for nitrogen. Yet, this hypothesis is highly speculative at present and awaits further experimental data.
Figure 2.—
Analysis of Δtor1 Δtor2 double mutants. (A) Schematic of the opposite phenotypes of Δtor1 and Δtor2 mutations. (B) Terminal phenotype of Δtor1 Δtor2 cells expressing the nmt1-tor2+ construct (TA426) in the presence or absence of thiamine. (C) FACS analysis. Δtor2 or Δtor1 Δtor2 mutant cells expressing nmt1-tor2+ (TA313 and TA426, respectively) were grown to midlog phase and thiamine was added.
To examine the genetic relationship between tor1+ and tor2+, we constructed a double-mutant strain Δtor1 Δtor2 expressing the conditional nmt1-tor2+ allele. When shifted to repressing conditions, Δtor1 Δtor2 cells divided for three to four divisions before growth arrest. The double-mutant cells did not reach the small cell size observed in repressed single tor2 mutants, although some cell size reduction occurred (Figure 2B). FACS analysis revealed that tor1 tor2 double mutants arrested at G2 with only a small fraction of cells at G1 (Figure 2C). Thus, the terminal phenotype of Δtor1 Δtor2 is intermediate compared with that of each of the single mutants. One possibility is that the tor2 mutation is epistatic to tor1, but since the initial cell length of Δtor1 cells is slightly longer than wild type, the Δtor1 Δtor2 cells reach a longer terminal size. Alternatively, Tor1 and Tor2 may regulate cell size and cell cycle progression independently.
We also examined the ability of Δtor1 Δtor2 nmt1-tor2+ cells to enter the sexual development pathway upon depletion of Tor2. Upon shutoff of tor2+ expression, we found no mating activity either in rich or in minimal medium. Thus, Tor1 acts in regulating sexual development either downstream of Tor2 or independently.
Taken together, our analysis of the Δtor1 Δtor2 double mutant reveals complex interactions, which may only partially support the suggestion that tor1+ loss of function leads to overactivation of Tor2. It suggests that Tor1 exerts at least some of its functions independently of Tor2.
Tor1 and the Tsc1–Tsc2 complex regulate leucine uptake and transcription of amino acid permeases by two separate mechanisms:
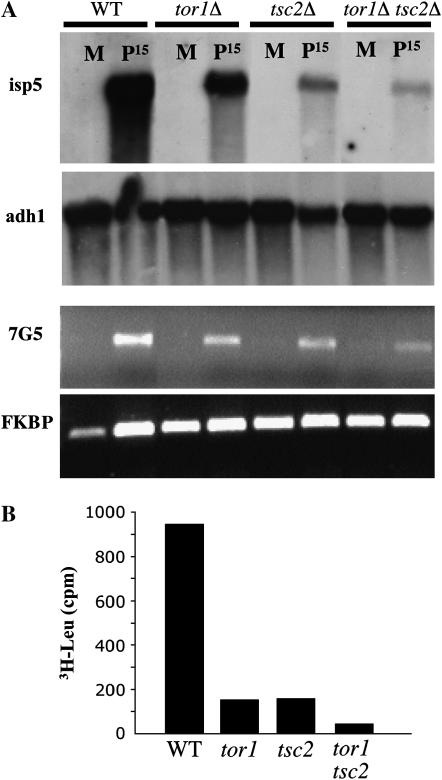
It has previously been shown that Tsc1/2 and Tor1 are both required for efficient amino acid uptake and expression of three different amino acid permeases: isp5+, 7G5.06, and c869.10 (Van Slegtenhorst et al. 2004; Weisman et al. 2005). To explore the genetic relationships between tor1+ and tsc1/2+ we determined the expression of these permeases in the double mutants tor1 tsc1 and tor1 tsc2. Since transcription of isp5+ is almost undetectable by Northern blotting in the presence of ammonia, we compared isp5+ transcription 15 min after a shift to proline-based medium. As expected, expression of isp5+ is reduced in the single tsc2 or tor1 mutants. Further reduction is detected in the double mutant (Figure 3A), suggesting that Tor1 and Tsc2 regulate isp5+ transcription by parallel mechanisms. Similar results were found for Δtsc1 and Δtor1 Δtsc1, respectively (data not shown). Using semiquantitative RT–PCR analysis we also observed an additive decrease in the expression of the 7G5.06 permease in the double mutant tor1 tsc2 compared to the single mutants (Figure 3A). Finally, we directly measured the uptake of radioactive leucine. Figure 3B shows that leucine uptake is reduced in the double mutants, compared to single mutants, in accord with the additive effects on transcription of amino acid permeases.
Figure 3.—
Additive effects of Δtor1 and Δtsc2 mutations. (A) Wild-type (TA001), Δtor1 (TA390), Δtsc2 (TA450), and Δtor1 Δtsc2 (TA459) cells were grown to midlog phase in EMM (M) medium, washed, resuspended in proline (P) medium, and further incubated for 15 min at 30°. RNA was extracted and subjected to Northern blot (top two sections) or semiquantitative RT–PCR analyses (bottom two sections). Northern blots were probed with isp5+ and adh1+ (loading control). For RT–PCR analysis, primers were specific for the amino acid permease 7G5.06 or FKBP12 (control). (B) Δtor1 (TA390), Δtsc2 (TA450), and Δtor1 Δtsc2 (TA459) cells were grown to midlog phase in EMM medium. Incorporation of l-3H-leucine was measured after 6 min using the protocol detailed in materials and methods.
Either Tor1 or Tsc1–Tsc2 is required for growth when the nitrogen source in the medium is proline:
The defect in leucine uptake in Δtor1 cells or rapamycin-treated cells can be suppressed by replacing the rich nitrogen source in the medium (ammonia) by a poorer source, such as proline (Weisman et al. 2005). This suppression likely involves transcriptional induction of amino acid permeases in the presence of a poor nitrogen source such as proline (Weisman et al. 2005). Proline also suppresses the amino acid uptake defect of tsc1 or tsc2 mutants, enabling auxotrophic tsc1 tsc2 mutants to grow on proline, but not on ammonia plates that are normally supplemented with the required amino acids (Figure 4A). However, unexpectedly, the growth of prototrophic tsc1 or tsc2 mutants was highly sensitive to rapamycin on proline plates, but not on ammonia plates (Figure 4B). Rapamycin inhibited the growth of tsc1/2 mutants on proline plates, but not when the nitrogen source in the medium was glutamic acid, a different amino acid that is more easily metaboilized by yeasts (Figure 4B). Thus, sensitivity of tsc1/2 to rapamycin is specific to a poor nitrogen source and may represent inability to metabolize proline, rather then inability to uptake amino acids.
Figure 4.—
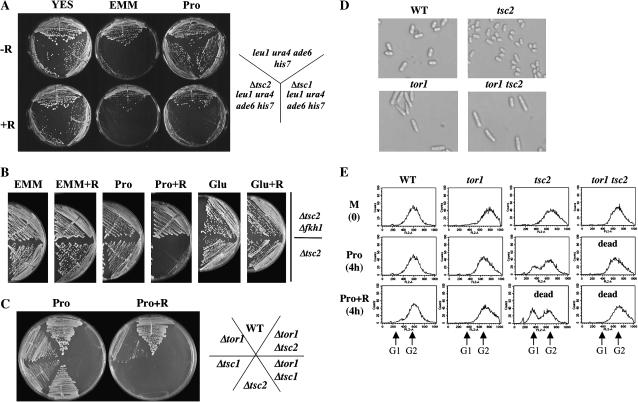
Δtsc2 mutants are sensitive to rapamycin on medium containing a poor nitrogen source. (A) Δtsc1 or Δtsc2 mutants (TA419 and TA418, respectively) were streaked onto rich yeast extract (YE) or minimal plates containing ammonia (EMM) or proline (Pro) as the sole nitrogen source. Addition of rapamycin to the proline medium inhibited the growth on proline but not on rich medium. (B) Sensitivity to rapamycin on proline medium requires FKBP12. Prototrophic Δtsc2 mutants (TA450) or Δtsc2 Δfkh1 double mutants (TA468) were streaked on yeast extract (YE), proline (Pro), or glutamic acid (Glu) plates in the presence or the absence of 0.1 μg/ml rapamycin. Plates were incubated for 4 days at 30°. (C) Δtsc2 mutant cells were unable to grow on proline plates when combined with the Δtor1 mutation. The indicated strains, corresponding to TA001, TA390, TA449, TA481, TA450, and TA459, were streaked on proline plates in the presence or the absence of 0.1 μg/ml rapamycin. (D) Deletion of tor1+ reverses the small cell size and G1 accumulation of Δtsc2 mutants. Wild-type (TA001), Δtor1 (TA390), Δtsc2 (TA450), and Δtor1 Δtsc2 (TA459) cells were grown to late log phase in minimal EMM media at 30° before being photographed. Deletion of tor1+ also reverses the small cell size and G1 accumulation of Δtsc1mutants (data not shown). (E) FACS analysis of the strains indicated in D. Cells were grown to midlog phase in EMM (M) media and then shifted to either proline (Pro) or proline with 0.1 mg/ml rapamycin (Pro + R).
tsc2 mutants that also carried a mutation in the S. pombe FKBP12 gene, fkh1+, were completely resistant to growth inhibition by rapamycin on proline (Figure 4B). Similar results were seen for the tsc1 fkh1 double mutant (data not shown). The FKBP12-dependent rapamycin sensitivity of tsc1/2 suggests inhibition of a TOR-dependent function. We next examined the possibility that rapamycin may inhibit a Tor1-dependent function, by examining the ability of tor1 tsc1 or tor1 tsc2 mutants to grow on proline. As seen in Figure 4C the double mutants tor1 tsc1 or tor1 tsc2 failed to grow on proline plates, suggesting that Tsc1/2 and Tor1 act separately to regulate growth in the presence of a poor nitrogen source and that this function of Tor1 is sensitive to rapamycin.
The small cell size phenotype of tsc1 or tsc2 cells is reversed by tor1 disruption:
While both Tor1 and Tsc1/2 positively regulate amino acid uptake, cell size appears to be oppositely regulated by Tor1 and Tsc1/2. Δtor1 cells are elongated (Weisman and Choder 2001; Matsuo et al. 2003); in contrast, we noted that single tsc1 or tsc2 mutants are slightly smaller than isogenic wild-type cells (Figure 4D). We examined cell length of the double mutants tor1 tsc1 or tor1 tsc2 and found that the tor1 mutation reverses the short cell size phenotype of tsc1/2 mutants (Figure 4D, data are shown for the tsc2 mutation only). Thus, with respect to cell size, Tor1 may act downstream or independently of Tsc1/2.
When cells are shifted to less favorable growth conditions, such as poor nitrogen sources, mitosis is advanced and cells divide at a smaller size (Fantes and Nurse 1977; Young and Fantes 1987). Shift of tsc2 mutants from ammonia to proline medium (nutritional downshift) resulted in further cell size shortening and in the appearance of a small G1 peak (Figure 4E). This G1 peak likely reflects the small cell size of the tsc1/2 mutants and thus the activation of a cell-size G1/S checkpoint. Shift to proline in the presence of rapamycin resulted in further shortening of cell size and accumulation of even more cells at the G1 phase (Figure 4E). Similar data were observed for tsc1 mutants (data not shown). These findings indicate that tsc1/2 mutants still respond to shift to proline by cell-size shortening and that loss of function of tsc1/2 is additive to the effect of rapamycin. The target of rapamycin that causes this cell size shortening is yet unclear. Since loss of function of Tor2, but not of Tor1, leads to short cell size, it is possible that Tor2 inactivation leads to rapamycin-induced cell-size shortening. Growth arrest of tsc2 mutants in proline medium occurs irrespective of their terminal cell size, since tsc1/2 mutants die as small cells on proline in the presence of rapamycin, but as elongated cells when combined with the tor1 mutation.
Rapamycin treatment, like loss of function of tsc1 or tsc2, confers thialysine and canavanine resistance:
One of the more detailed analyzed characteristics of tsc mutants is their inefficient amino acid uptake, which leads to canavanine resistance (Van Slegtenhorst et al. 2004; Nakase et al. 2006). Since rapamycin treatment of wild-type cells leads to inefficient leucine uptake (Weisman et al. 2005), we examined their sensitivity in the presence of rapamycin to canavanine and thialysine, the later a toxic analog of lysine. We found that treatment with rapamycin results in resistance to these drugs, similar to the phenotype caused by deletion of tsc1/2 (Figure 5A). The rapamycin-induced resistance to canavanine or thialysine is abolished in cells carrying disruption of the FKBP12 gene (Figure 5A and data not shown), suggesting that rapamycin acts via inhibition of a TOR-dependent function. Either the FRB of Tor1 or that of Tor2 can bind the FKBP12-rapamycin complex, as we have shown by two-hybrid assays (Weisman 2004). However, as overexpression of Tor2 leads to canavanine resistance (Figure 1E), it is unlikely that the effect of rapamycin is due to direct inhibition of Tor2. A plausible mechanism to explain this effect of rapamycin would be the inhibition of a Tor1-dependent function. However, we found that tor1 mutants do not show resistance to canavanine and are slightly more sensitive to thialysine compared to wild-type cells (Figure 5B). Using higher concentrations of canavanine or a different incubation time we also observed that tor1 mutants are more sensitive to canavanine compared to wild-type cells (data not shown). This may be indicative of an increase in uptake of the toxic analogs in tor1 mutants that differs from the previously described uptake of leucine, histidine, or uracil. Since tor1 mutants are highly sensitive to two different toxic amino acid analogs, we prefer an alternative explanation in which the sensitivity of tor1 mutants to canavanine or thialysine reflects translational or protein folding defects.
At present the target of rapamycin in inducing canavanine or thialysine resistance is unclear. The possibility that rapamycin completely inhibits Tor1 activity is unlikely, since Δtor1 mutant do not show canavanine or thialysine resistance. Yet, it is possible that rapamycin inhibits certain Tor1-dependent activities, leading to reduced amino acid uptake. As noted above, Tor1 may negatively regulate Tor2, via a negative feedback loop, and thus its inhibition by rapamycin may remove the negative feedback loop, leading to overactivation of Tor2 and, thus, a decrease in amino acid uptake (Figure 5C).
DISCUSSION
Disruption of TOR activity in budding yeast, nematodes, Drosophila, mice, and human cells leads to features that are characteristic of starved cells. These starvation-like phenotypes have suggested that TOR plays a conserved role in regulating cell growth in response to nutrient availability. So far, studies of TOR signaling in S. pombe did not fit into such a model since the only characterized TOR homolog in fission yeast, Tor1, is mainly required for starvation and stress responses and its absence results in inability to respond to starvation, rather then exhibiting a starvation-like phenotype. In contrast with Tor1 loss of function, we show here that disruption of tor2+ results in a phenotype that highly resembles nitrogen starvation. Cells depleted of Tor2 arrest their growth at the G1 phase of the cell cycle as small and rounded cells, induce expression of nitrogen-starvation-specific genes, and are derepressed for sexual development. Most importantly, this phenotype highly resembles that of disruption of rhb1+, suggesting that the link between Rheb and TOR signaling is conserved in evolution from S. pombe to human. Previously, a physical interaction was described between Tor2 and hyperactivated Rhb1 in fission yeast (Urano et al. 2005). We suggest that Tor2 lies downstream of Rhb1. Yet, since overexpression of tor2+ did not suppress rhb1+ deletion (data not shown), final proof awaits isolation of hyperactivated Tor2. While this article was under review two articles (Alvarez and Moreno 2006; Uritani et al. 2006) were published that support our findings. Both demonstrate that Tor2 depletion leads to a phenotype of nitrogen-starved cells, including arrest as small rounded cells, induction of nitrogen-specific genes, such as isp6+, and entrance into sexual development under rich medium conditions.
The always starved for nitrogen phenotype of tor2 mutants is in striking contrast to the never starved for nitrogen phenotype of tor1cells (Figure 2A). Since Tor1 and Tor2 appear to regulate similar processes in an opposite fashion, it is tempting to speculate that their functions are interconnected. One possibility is that Tor1 negatively inhibits Tor2; thus in the absence of Tor1, Tor2 may become hyperactivated, signaling availability of nitrogen even in the absence of a nitrogen source. Yet other inhibitory links between the two TOR-dependent pathways may explain these opposite effects. Tor1 is involved in additional, Tor2-independent processes. For example, stress response and acquisition of long-term viability in stationary phase are two functions regulated by Tor1, but not by Tor2 (Figure 5C).
While Tor2 is likely to be positively regulated by Rhb1, Tor1 appears to act in parallel with Tsc1/2 with respect to amino acid uptake, regulation of amino acid permeases, and in regulating growth when proline is used as the nitrogen source. Most recently, Yang et al. (2006) found that the Drosophila Rheb and mammalian TSC1/2 have opposite effects on TORC1 and TORC2. These opposite effects are reminiscent of the opposite effects of TSC1/2–Rhb1 on Tor1 and Tor2. Our results show additive effects of the tor1 and tsc1/2 mutations: either tor1+ or tsc1/2+ is required for growth on poor nitrogen source and downregulation of amino acid permeases or leucine uptake is more pronounced in the double mutant tsc1 tor1 compared to the relevant single mutants. These findings argue against a direct effect of Tsc1/2–Rhb1 on Tor1. Rather, Tsc1/2–Rhb1 and Tor1 may act in two parallel pathways.
In many organisms, treatment with rapamycin results in TOR loss of function, often leading to a G1 cell cycle arrest and to a phenotype of starved cells. In contrast, rapamycin does not affect wild-type S. pombe cells grown on rich medium. We have previously shown that auxotrophic S. pombe strains are sensitive to rapamycin due to inhibition of a Tor1-dependent function that positively regulates amino acid uptake. Here we identify tsc1/2 mutants as sensitive to rapamycin when grown on proline medium. This is clearly unexpected if Tsc1/2 in S. pombe were acting solely as negative regulators of TOR signaling. Accumulation of data now suggests that the phenotype of loss of function of tsc1/2+ is very similar to treatment with rapamycin. Both tsc1/2 mutants and rapamycin treatment result in partial sterility, decrease in leucine uptake, downregulation of permeases, resistance to canavanine, small cell size, and accumulation in G1 (Weisman et al. 1997, 2005; Matsumoto et al. 2002; Van Slegtenhorst et al. 2004; this study). Tsc1/2 may act in parallel with a Tor1-dependent rapamycin-sensitive function. Yet, cells deleted for tor1+ are still sensitive to rapamycin in an FKBP12-dependent manner (Weisman 2004). Thus, whether certain Tor2 functions are also inhibited by rapamycin is as yet an open question. Our findings that rapamycin treatment can lead to cellular effects that mimic loss of function of Tsc1/2, rather than reversing the effects of mutations in Tsc1/2, draw attention to the complexity of the TOR signaling pathway and stress the need for more detailed information on the link between TSC and TOR.
Acknowledgments
We thank Marjon van Slegtenhorst for tsc1 and tsc2 mutations and for very helpful discussions during the preparation of this work. We thank Fuyuhiko Tamanoi for rhb1 mutants and rhb1+ plasmids. This work was supported by grants to R.W. by the Israel Science Foundation (397/03-16.2) and by the Association for International Cancer Research (02-074).
References
- Alvarez, B., and S. Moreno, 2006. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119: 4475–4485. [DOI] [PubMed] [Google Scholar]
- Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite et al., 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7: 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., and M. N. Hall, 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Booher, R., and D. Beach, 1986. Site-specific mutagenesis of cdc2+, a cell cycle control gene of the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 6: 3523–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como, C. J., and K. T. Arndt, 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10: 1904–1916. [DOI] [PubMed] [Google Scholar]
- Fantes, P., and P. Nurse, 1977. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp. Cell Res. 107: 377–386. [DOI] [PubMed] [Google Scholar]
- Gangloff, Y. G., M. Mueller, S. G. Dann, P. Svoboda, M. Sticker et al., 2004. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell. Biol. 24: 9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L. S., G. M. Findlay, A. Gray, T. Tolkacheva, S. Wigfield et al., 2004. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, N., and N. Sonenberg, 2004. Upstream and downstream of mTOR. Genes Dev. 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S., 2005. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33: 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, E., R. Loewith, A. Schmidt, S. Lin, M. A. Ruegg et al., 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6: 1122–1128. [DOI] [PubMed] [Google Scholar]
- Kawai, M., A. Nakashima, M. Ueno, T. Ushimaru, K. Aiba et al., 2001. Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39: 166–174. [DOI] [PubMed] [Google Scholar]
- Kim, D. H., D. Sarbassov dos, S. M. Ali, R. R. Latek, K. V. Guntur et al., 2003. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11: 895–904. [DOI] [PubMed] [Google Scholar]
- Li, Y., M. N. Corradetti, K. Inoki and K. L. Guan, 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29: 32–38. [DOI] [PubMed] [Google Scholar]
- Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo et al., 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Long, X., C. Spycher, Z. S. Han, A. M. Rose, F. Muller et al., 2002. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 12: 1448–1461. [DOI] [PubMed] [Google Scholar]
- Mach, K. E., K. A. Furge and C. F. Albright, 2000. Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, B. D., and L. C. Cantley, 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28: 573–576. [DOI] [PubMed] [Google Scholar]
- Matsumoto, S., A. Bandyopadhyay, D. J. Kwiatkowski, U. Maitra and T. Matsumoto, 2002. Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics 161: 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, T., Y. Kubo, Y. Watanabe and M. Yamamoto, 2003. Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22: 3073–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Murakami, M., T. Ichisaka, M. Maeda, N. Oshiro, K. Hara et al., 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24: 6710–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase, Y., K. Fukuda, Y. Chikashige, C. Tsutsumi, D. Morita et al., 2006. A defect in protein farnesylation suppresses a loss of Schizosaccharomyces pombe tsc2+, a homolog of the human gene predisposing tuberous sclerosis complex. Genetics 173: 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., and Y. Ohsumi, 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273: 3963–3966. [DOI] [PubMed] [Google Scholar]
- Oldham, S., J. Montagne, T. Radimerski, G. Thomas and E. Hafen, 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14: 2689–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice, H. L., 1992. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 20: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov, D. D., S. M. Ali, D. H. Kim, D. A. Guertin, R. R. Latek et al., 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14: 1296–1302. [DOI] [PubMed] [Google Scholar]
- Sarbassov, D. D., D. A. Guertin, S. M. Ali and D. M. Sabatini, 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101. [DOI] [PubMed] [Google Scholar]
- Shah, O. J., Z. Wang and T. Hunter, 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14: 1650–1656. [DOI] [PubMed] [Google Scholar]
- Snaith, H. A., and S. L. Forsburg, 1999. Rereplication phenomenon in fission yeast requires MCM proteins and other S phase genes. Genetics 152: 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler, S., E. Warbrick, S. Prochnik, S. Mackie and P. Fantes, 1996. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109(7): 1927–1935. [DOI] [PubMed] [Google Scholar]
- Stiefel, J., L. Wang, D. A. Kelly, R. T. Janoo, J. Seitz et al., 2004. Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3: 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee, A. R., and J. Blenis, 2005. mTOR, translational control and human disease. Semin. Cell Dev. Biol. 16: 29–37. [DOI] [PubMed] [Google Scholar]
- Um, S. H., F. Frigerio, M. Watanabe, F. Picard, M. Joaquin et al., 2004. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200–205. [DOI] [PubMed] [Google Scholar]
- Urano, J., M. J. Comiso, L. Guo, P. J. Aspuria, R. Deniskin et al., 2005. Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol. Microbiol. 58: 1074–1086. [DOI] [PubMed] [Google Scholar]
- Uritani, M., H. Hidaka, Y. Hotta, M. Ueno, T. Ushimaru et al., 2006. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells 11: 1367–1379. [DOI] [PubMed] [Google Scholar]
- Van Slegtenhorst, M., E. Carr, R. Stoyanova, W. Kruger and E. P. Henske, 2004. Tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279: 12706–12713. [DOI] [PubMed] [Google Scholar]
- Weisman, R., 2004. The fission yeast TOR proteins and the rapamycin response: an unexpected tale. Curr. Top. Microbiol. Immunol. 279: 85–95. [DOI] [PubMed] [Google Scholar]
- Weisman, R., and M. Choder, 2001. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276: 7027–7032. [DOI] [PubMed] [Google Scholar]
- Weisman, R., M. Choder and Y. Koltin, 1997. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J. Bacteriol. 179: 6325–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman, R., I. Roitburg, T. Nahari and M. Kupiec, 2005. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics 169: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger, S., R. Loewith and M. N. Hall, 2006. TOR signaling in growth and metabolism. Cell 124: 471–484. [DOI] [PubMed] [Google Scholar]
- Yang, Q., K. Inoki, E. Kim and K. L. Guan, 2006. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. USA 103: 6811–6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., A. P. Tabancay, Jr., J. Urano and F. Tamanoi, 2001. Failure to farnesylate Rheb protein contributes to the enrichment of G0/G1 phase cells in the Schizosaccharomyces pombe farnesyltransferase mutant. Mol. Microbiol. 41: 1339–1347. [DOI] [PubMed] [Google Scholar]
- Young, P. G., and P. A. Fantes, 1987. Schizosaccharomyces pombe mutants affected in their division response to starvation. J. Cell Sci. 88(3): 295–304. [DOI] [PubMed] [Google Scholar]
- Zaragoza, D., A. Ghavidel, J. Heitman and M. C. Schultz, 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18: 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard and T. P. Neufeld, 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14: 2712–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]