Abstract
Combined with a system for identifying each of the chromosomes in a genome, visualizing the location of individual genetic loci by fluorescence in situ hybridization (FISH) would aid in assembling physical and genetic maps. Previously, large genomic clones have been successfully used as FISH probes onto somatic chromosomes but this approach is complicated in species with abundant repetitive elements. In this study, repeat-free portions of sequences that were anchored to particular chromosomes including genes, gene clusters, large cDNAs, and portions of BACs obtained from public databases were used to label the corresponding physical location using FISH. A collection of probes that includes at least one marker on each chromosome in the maize complement was assembled, allowing a small-target karyotyping system to be developed. This set provides the foundation onto which additional loci could be added to strengthen further the ability to perform chromosomal identification in maize and its relatives. The probes were demonstrated to produce signals in several wild relatives of maize, including Zea luxurians, Z. diploperennis, and Tripsacum dactyloides.
COMBINED with the ability to detect individual loci, fluorescence in situ hybridization (FISH) identification of each of the chromosomes in somatic preparations has proven useful for cytological mapping of transgene insertions (Kato et al. 2006), gene clusters (Kato et al. 2006; Valdivia et al. 2007), and active transposon insertions (Yu et al. 2007). Maize somatic chromosomes have been identified using a cocktail of FISH probes consisting of labeled repetitive elements that are found in large arrays at specific locations in the genome (Kato et al. 2004). Although effective in all tested inbred lines, the application of the repetitive element cocktail for somatic karyotyping has some limitations. The presence and copy number of repetitive elements varies among maize lines (Rivin et al. 1986; Kato et al. 2004; Lamb and Birchler 2006) and related species (Lamb and Birchler 2006). Therefore, the hybridization pattern must be determined for each line. In contrast, gene sequences and order are generally more conserved in maize and its relatives (see below). A karyotyping system based on features such as genes would allow chromosomes to be identified without prior line characterization.
In plant species with small gene-rich genomes, such as Arabidopsis thaliana (Koornneef et al. 2003), rice (Jiang et al. 1995), and sorghum (Kim et al. 2002, 2005), detection of specific loci can be accomplished by FISH with large genomic clones as probes. Unfortunately, the genomes of many species of agronomic and scientific interest contain families of abundant repetitive elements (Bennetzen and Kellogg 1997). As a result, any clone that includes a copy of a repetitive element will hybridize across the genome, making it unsuitable as a FISH probe (Lamb and Birchler 2006; Lamb et al. 2007). In some cases, addition of competitor DNA that blocks probe hybridization to repetitive sequences allows large genomic clones to be detected. This approach, called competitive in situ suppression, was used successfully to detect the position of cosmid clones on maize pachytene and somatic chromosomes (Sadder et al. 2000; Sadder and Weber 2001, 2002). Because large genomic clones of maize differ in type and content of repetitive elements, the suppression conditions for each clone must be determined empirically. One solution is the use of sorghum genomic clones as FISH probes (Zwick et al. 1998; Koumbaris and Bass 2003). This approach takes advantage of the conservation of gene sequences between the two species and the divergence of their repetitive elements. However, to find suitable sorghum BACs that correspond to particular maize sequences, a sorghum genomic BAC library must be screened and this approach might be complicated by rearrangements between the two taxa.
Recent improvements to FISH techniques allow the detection of very small maize genomic targets (Kato et al. 2006; Wang et al. 2006; Yu et al. 2007). Because unique sequences can be chosen as targets, this method provides a straightforward means for circumventing the problem of abundant repetitive elements in plant genomes.
In this study, we describe several approaches for identifying sequences to be used as FISH probes that label specific maize loci. A set of probes was assembled, including at least one for each maize chromosome. This collection serves as the foundation for building an increasingly precise collection of probes for karyotyping studies on chromosomes of maize and its relatives and illustrates the means for producing a custom-made set of probes for specific regions of the genome. Using a subset of this collection as a karyotyping cocktail, it is possible to identify each maize chromosome and chromosomes in related Zea and Tripsacum species.
MATERIALS AND METHODS
Plant materials:
Maize × Zea diploperennis hybrid seed (NC300 × PI 441932) was provided by J. Holland (North Carolina State University). Maize × Z. luxurians (ig1 × Z. luxurians, 00w-268-2X12) hybrid seed was provided by J. Doebley (University of Wisconsin). Maize × Tripsacum dactyloides seed was obtained by applying Tripsacum pollen onto popcorn (Supergold). The “tri-species” hybrid was produced by W. Galinat and has been described previously (Lamb and Birchler 2006). It was obtained by applying pollen from T. dactyloides onto maize silks, doubling the chromosome number of the resultant plants with colchicine, and then crossing with Z. diploperennis. Subsequently, maize chromosome 2 was lost during vegetative propagation. As a result, the current plant has a haploid contribution from Z. diploperennis (n = 10), T. dactyloides (n = 18), and nine maize chromosomes (1 and 3–10).
FISH technique:
Somatic chromosome preparations (Yu et al. 2006), direct probe labeling by nick translation using a high concentration of DNA polymerase (Kato et al. 2006; Lamb et al. 2006; Yu et al. 2007), and the FISH procedure (Kato et al. 2004, 2006) were as described previously. A drop of Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA) and a glass coverslip were added to each slide to preserve and counterstain chromosome preparations. Images were captured with an Olympus BX61 microscope using Applied Spectral Imaging (ASI) software and the cooled charge-coupled device camera Cool-1300QS. Images were then processed using the “sharpen” feature of the ASI software. The single-locus signals presented as gray values were only minimally digitally adjusted (using the curves feature of Photoshop), except for myo1 where background was subtracted by overlaying a severely blurred copy of the image using the “difference” function of Photoshop as previously described (Kato et al. 2004). Merged RGB images were digitally enhanced to clearly show the position of signals on the chromosomes.
In interspecies hybrids, chromosomes of Z. mays, Z. diploperennis, and T. dactyloides were identified using retroelement FISH probes that differentially paint chromosomes from different species (Lamb and Birchler 2006). The Grande retroelement probe strongly labels maize chromosomes but hybridizes with only intermediate intensity to Z. diploperennis chromosomes. Retroelement probe TC#25 specifically hybridizes to Tripsacum chromosomes (Lamb and Birchler 2006). Distal knob positions are characteristic of Z. luxurians chromosomes, allowing them to be distinguished from maize chromosomes in interspecies hybrids (Lamb et al. 2007).
Probe development:
To develop a collection of large single-locus FISH probes that includes at least one representative on each chromosome, targets needed to be identified on chromosomes 1, 5, 7, 8, and 9 (useful probes for the other chromosomes were already available). BAC sequence AF448416 (106,186 bp) spanning the bronze1 genomic region was selected for developing a probe for chromosome 9 (Fu and Dooner 2002; Brunner et al. 2005). For chromosomes 5 and 8, the EMBL database was searched for relevant sequences. Two Z. mays gene sequences were arbitrarily selected for chromosome 5 probe development: somatic embryogenesis receptor-like kinase 2 (Serk2, 5484 bp, AJ400869) and fatty aldehyde dehydrogenase 1 (rf2e1, 4739 bp, AY374447). BAC sequence AC157487 was selected for chromosome 8 probe development. Repetitive elements in these sequences were identified using the RepeatMasker program (Smit et al. 2004). Unmasked regions of the sequences were compared against the NCBI database using BLASTn (Altschul et al. 1990). Primers were designed to PCR amplify regions with homology to plant cDNAs or mRNAs (Table 1) using Oligo 6.31 Primer Analysis software (Molecular Biology Insights, http://www.oligo.net/). PCR products of the expected size were cut and eluted from agarose gels (Promega, Madison, WI, Wizard SV gel and PCR clean-up system), reamplified if necessary, and tested individually as FISH probes (Figure 2). Probes that showed little background were cloned (Promega pGEM-T vector system) and sequenced to confirm their identity to the GenBank sequence. Multiple PCR products spanning each genomic sequence were combined to produce the final FISH probe.
TABLE 1.
PCR probe production
| Chromosome arm | Probe name | Selected PCR product length (bp) | Primers 5′–3′ | Template | Sequence accession no. |
|---|---|---|---|---|---|
| 1S | dek1 | 6861 | GGA CAC CAC GGA GTT GTT TT TGC GTT AAC AGG AAC GAC AG | Whole RNA and mRNA from leaves, Oh43 | AY061806 |
| 2S/10L | acc1/acc2 | 7090 | CTC AAA GGC CTT GCC ACT AC GCA CCA GGT CCA AAA GAA AA | mRNA from leaves, Oh43 | AY312171 |
| 3L | myo1 | 2674 | ACC ATT TCT GCC CTC CGT CCC CTC TCA ACT TAT TTT CAC TGC TGC TGC TTC CTG CCT CAT | mRNA from leaves, Oh43 | AF104924 |
| 2489 | TGA GGC AGG AAG CAG CAG CAG TGA AAA TAC GCA TCC GGC CCC TTT TCA CTA CAG | ||||
| 4 | Cent4-279 | 279 | CCA AAG ATC ATA AAG AAT TAA GGG GGT TTG ATG TTT ACG TTG GAC | Plasmid A8, Bluescript | AF242891 |
| 5L | serk2 | 5460 | GTT GAC TTT GAG GTG CGA TT CAC AAC CTC AGC GTA ACC AAC | Genomic DNA B73 | AJ400869 |
| 5L | rf2e1 | 1670 | GGA GGA GAT CTT CGA CGT G AAC AGG TGG TAA CGC AGA TAG | Genomic DNA B73 | AY374447 |
| 1852 | GCC AAC ATA CCC AAG TAC CT ATG TTT GTC AAT TTC CGT ACA | ||||
| 7S | α-zeinB | 770 | CGG CAC GAG GCA ACA TAG AAA GT TAA AAG AGG GCA CCA CCA ATG ATG | Genomic DNA B73 | AF546188 |
| 8L | BAC8L | 3125 | CCA CTC CAC GGC TCC AAT AG GTC CAA GGC TAA TGC CTC TTC | Genomic DNA B73 | AC157487 |
| 2179 | TAG CAT ATT CGG TCC TG GAT GAT TTT GCA GCA CTA GAG | ||||
| 3353 | CCG AGT ATT TCC CAA CAT CTA ACG TCT TCC CGA TCT C | ||||
| 9S | BAC9S | 3480 | TCG ATC ATC ATC TCC TGC GAA TTT C ATT GTC ACG GGA TCT GGT CTA AAC | Genomic DNA B73 | AF448416 |
| 1636 | CTA TGT TCT TTC CTT CTT GCG TTA T GCG ATT TCT TCC TCA CCT ACT CC | ||||
| 1899 | TCT TGA AAT TTA GAC TTC GTG TGC C GTG ATT TGT TTT AGC TTG CGT GT | ||||
| 3444 | ACA GGC AAG AGC ATG AAC GTG TGC GGT TGT GCG TAT GA | ||||
| 1869 | GGG CAA CTT TGT TAC ACT CAC GAA C TTC CAC GCT TCA ATT TCC CTA CCT |
Figure 2.—
Development of unique probes from BAC sequences. (A) Chromosome 8L anchored BAC AC157487 (136.9 kb) after RepeatMasker analysis: four long unique BAC regions were selected for primer design. (B) Seven PCR products were amplified and used as FISH probes separately. Three PCR products that showed low or no background were selected and combined as one probe.
A search algorithm that identifies mRNAs of a given size from sequences deposited in GenBank was used to find candidate sequences for FISH probe development. This algorithm filters GenBank entries by species, type, and size and is provided in the form of a PERL script in the supplemental materials at http://www.genetics.org/supplemental/. Maize sequences that are annotated as cDNAs or mRNAs and that are >6000 bp were manually examined for those that were previously mapped. The dek1 mRNA (AY061806, 7110 bp) was chosen for development of a chromosome 1 specific probe. Two additional long mRNAs, including one whose genomic position was unknown, were selected for use as FISH probes: the unconventional myosin heavy chain (myo1) (5375 bp) and acetyl-coenzyme A carboxylase (acc1) (7324 bp). PCR primers were designed for reverse transcriptase–PCR reaction (Table 1).
The 19-kDa zein gene family is organized as clusters at multiple locations throughout the maize genome, including a subfamily cluster on chromosome 7S (Song and Messing 2002). PCR products specific to the B subfamily were amplified using primers described by Song and Messing (2002). The product was cloned, yielding plasmid pAZB6-2, and sequenced to confirm its identity. Using the BLASTn algorithm (Altschul et al. 1990), the sequence was found to be identical to one of the six 19-kDa zein genes found in the 203,363-bp sequenced region located on chromosome 7S (GenBank accession AF546188).
PCR conditions:
B73 genomic DNA was extracted from leaves with the DNeasy plant mini kit from QIAGEN (Chatsworth, CA) and used as a template for all PCR reactions. Table 1 lists all single-locus probes and primers used. For amplifying 1- to 7-kb products, JumpStart REDAccuTaq LA DNA polymerase from Sigma (St. Louis) was used. PCR conditions were as recommended by the manufacturer: an annealing temperature of 62°, an extension temperature of 68°, and an extension time of 1 min for each 1000 bp. Plasmid inserts were amplified with standard primers (M13F/M13R, T3/T7, or T7/Sp6) and standard conditions using JumpStart REDTaq ReadyMix PCR reaction mix (Sigma). For reverse transcriptase PCR, template RNA was extracted from leaves or immature endosperm tissue using Trizol reagent (Invitrogen, San Diego), and long cDNAs were amplified using Invitrogen's SuperScript III one-step RT–PCR system with Platinum Taq High Fidelity polymerase. To eliminate nucleolus organizer region (NOR) hybridization by cDNA-based FISH probes, the RNA template was enriched for poly(A) containing mRNA using Promega's PolyAtract mRNA isolation system IV.
Karyotyping:
Chromosomes from the B73, Oh43, and KYS inbred lines were identified using two different cocktails of repetitive element FISH probes. The first contains CentC and the TAG microsatellite labeled with AlexaFluor 488-5-dUTP (Figure 1, green) and the 180-bp knob repeat labeled with coumarin-5-dUTP (Figure 1, blue). An alternative cocktail consists of CentC and the subtelomeric repeat 4-12-1 labeled with AlexaFluor 488-5-dUTP and the 180-bp knob repeat labeled with coumarin-5-dUTP. Chromosome identification using this cocktail relies on FISH signal position as reported (Kato et al. 2004) as well as arm-length ratios, chromosome size, and position of the NOR constriction.
Figure 1.—
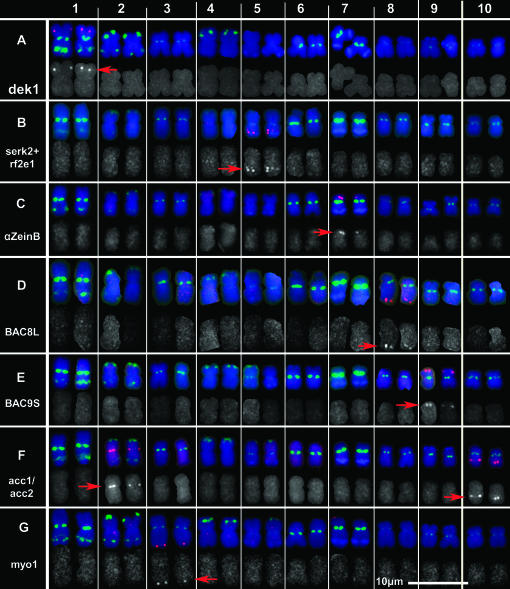
Small-target FISH on somatic chromosomes from maize inbred line B73. Somatic chromosomes from inbred line B73 were hybridized with small-target probes (red) and with repetitive element probes (CentC, TAG microsatellite, and the 180-bp knob repeat), which in combination with size and arm-length ratios, allow each chromosome to be identified. CentC and TAG microsatellite signals are green and the 180-bp knob repeat signals are blue. Chromosomes from individual preparations were electronically cut out and arranged in rows. In each row, the merged image is presented to show the chromosomal position of each small-target probe. Below the merged image, the gray values are displayed for the small-target signals as follows: (A) dek1, (B) serk2+rf2e1, (C) 19-kDa α-zeinB gene family, (D) BAC8L, (E) BAC9S, (F) acc1/acc2, and (G) myo1. Red arrows indicate the positions of the signals.
To identify each chromosome in the maize complement with single gene probes, a combination of sequences was used (Table 2). Some components of the probe cocktail were produced using gene clones provided by S. Chopra (p1-wr gene clones SA204, pWRG57, and pF2cDNA), S. Hulbert (rp1 and rp3 gene clones), E. Valdivia (cDNA clones of expB11 and expB9), Kato et al. (2004) [rDNA 5S (p2-3-3), clone CL569181], and B. Page (Cent 4 probe, clone A8, AF242891). To eliminate knob homologous sequences from the Cent4 clone A8, a short PCR (279-bp) product was amplified with primers (Table 1) designed after BLASTn analysis of sequence AF242891. The short PCR product without knob sequences was used as a Cent4 probe.
TABLE 2.
Karyotyping cocktail components
| Chromosome | Probe (signal color) | Length (bp) | Probe composition | Probe concentration (ng/μl) | Initial probe concentration (ng/μl) | Volume (μl) | Relative distance of probe from centromere (%) | FISH-labeled genomic region |
|---|---|---|---|---|---|---|---|---|
| 1S | dek1 (red) | 6,861 | Texas red-5-dUTP labeled PCR product | 14.0 | 200 | 0.6 | 59.4 ± 5.2 | Z. mays B73 calpain-like protein (dek1) gene |
| 1S | p1 (white) | 7,912 (6,242, 1,670) | Cy5-dUTP labeled 2 PCR products mixture | 15.0 | 250 | 0.3 + 0.3 | 56.3 ± 6.0 | Z. mays pericarp color 1 genes |
| 2L | 5S (p2-3-3)(green) | 700 | Alexa Fluor 488-5-dUTP labeled PCR product | 1.0 | 50 | 0.2 | 85.0a | Z. mays 5S ribosomal RNA gene DNA sequence 5S-2-3-3 |
| 3L | rp3 (red) | 4,000 | Texas red-5-dUTP labeled PCR product | 25.0 | 250 | 1 | 21.7 ± 4.2 | rp3 gene cluster |
| 4 | Cent4-279 (green) | 279 | Alexa Fluor 488-5-dUTP labeled PCR product | 10.0 | 200 | 0.5 | 0.0 | Centromeric 4 satellite (Cent4) |
| 5L | serk2+rf2e1 (green) | 8,982 | Alexa Fluor 488-5-dUTP labeled 3 PCR products mixture | 50.0 | 500 | 1.0 | 62.5 ± 7.2 | Z. mays serk2 gene for somatic embryogenesis receptor-like kinase 2, Z. mays fatty aldehyde dehydrogenase 1 gene |
| 5L | expB11 (red) | 1,000 | Texas red-5-dUTP labeled PCR product | 12.5 | 250 | 0.5 | 85.2 ± 2.9 | Z. mays expansin B11 gene cluster |
| 6S | NOR (green) | 700 | Alexa Fluor 488-5-dUTP labeled PCR product | 0.025 | 1.25 | 0.2 | NOR rDNA genes | |
| 7S | α-zeinB (red) | ∼770 | Texas red-5-dUTP labeled PCR product | 25.0 | 250 | 1.0 | 78.1 ± 5.8 | α-zeinB genes cluster |
| 8L | BAC8L (green) | 8,657 | Alexa Fluor 488-5-dUTP 3 PCR products mixture | 50.0 | 500 | 1.0 | 79.3 ± 2.6 | Z. mays BAC 136,932-bp sequence anchored to chromosome 8 |
| 9S | BAC9S (green) | 12,328 | Alexa Fluor 488-5-dUTP labeled 5 PCR products mixture | 24.0 | 300 | 0.7 | 64.4 ± 7.5 | Z. mays B73 chromosome 9S bz genomic region |
| 9L | expB9 (green) | 900 | Alexa Fluor 488-5-dUTP labeled PCR product | 30.0 | 200 | 1.5 | 51.4 ± 4.6 | Z. mays expansin B9 gene |
| 10S | rp1 (red) | 1,700 | Texas red-5-dUTP labeled PCR product | 25.0 | 250 | 1.0 | 74.0 ± 5.7 | rp1 gene cluster |
| Total probe volume per one preparation (μl): | 10 | |||||||
Li and Arumuganathan (2001).
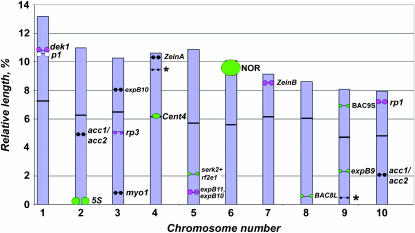
The idiogram was produced by calculating chromosome length, arm ratios, and relative probe distance (percentage of the arm from the centromere) using the computer application MicroMeasure version 3.3 (Reeves and Tear 2000). In addition to the probes used for karyotyping, other small-target probes displayed on the idiogram include expB10 (Valdivia et al. 2007) and the 19-kDa zein A gene (α-zeinA) (Kato et al. 2006). At least four chromosomes from the B73 inbred line were measured for each probe except α-zeinA and the 5S ribosomal gene cluster for which the position in different lines was determined from previous publications (Li and Arumuganathan 2001; Kato et al. 2006; Lamb and Birchler 2006).
RESULTS
Probe development:
Previous reports of single-locus FISH detection suggest that the minimum genomic target that can be detected using FISH in maize is ∼3000 bp (Kato et al. 2006; Wang et al. 2006; Yu et al. 2007). To develop probes that could be routinely used, several approaches identified genomic targets >6000 bp that would be free of repetitive elements and that would be expected to be readily detectable. Such targets included genes organized in clusters, large cDNAs, genes without repetitive elements in their introns, and pooled unique sequences from BACs.
Gene clusters:
Certain types of plant genes tend to be organized as large gene clusters. Because a single probe will hybridize to the entire group, such clusters make excellent FISH targets. The classic examples of tandem gene detection are the ribosomal genes that have been used in karyotyping for many different species, including maize (Li and Arumuganathan 2001; Kato et al. 2004). Additionally, genes that mediate maize disease resistance (Webb et al. 2002; Smith et al. 2004) or that encode storage (Woo et al. 2001) and cell-wall proteins (Wu et al. 2001) are found in large clusters and have been used as FISH probes in maize (Bauer and Birchler 2006; Kato et al. 2006; Lamb and Birchler 2006; Valdivia et al. 2007). The 19-kDa zein genes are present in clusters at several loci, including on chromosomes 4S and 7S (Song and Messing 2002). Because the 19-kDa zein subfamily A gene cluster on chromosome 4 is readily detectable using FISH (Kato et al. 2006), the B subfamily gene cluster was a promising candidate for a chromosome 7 marker. A 19-kDa zein B subfamily sequence was PCR amplified and cloned using subfamily-specific primers that have been previously described (Song and Messing 2002). This clone was used in subsequent PCR reactions to produce template for the FISH labeling reaction. The probe was called α-zeinB and produced a signal exclusively on 7S (Figure 1).
Single genes:
Most maize genes are >3 kb (Haberer et al. 2005), the minimum size that is routinely detectable by FISH. Therefore, individual genes are candidates as single-locus FISH probes although some will contain repetitive elements in their introns and be unsuitable. To develop probes for chromosome 5, PCR primer pairs were designed to amplify two genes located on 5L: the rf2e1 gene (4739 bp) and the serk2 gene (5484 bp) (Table 1). The rf2e1 gene was amplified in two fragments. The resulting PCR products produced FISH signals on 5L and showed no or low background. The PCR products from serk2 and rf2e1 genes were combined to produce a 5L probe, referred to as serk2+rf2e1 (Figure 1). On some chromosomes, two signals were produced, probably corresponding to the two genes (Figure 1 and supplemental Figure 1, A and B, at http://www.genetics.org/supplemental/). The serk2 gene is placed on the GRAMENE Z. mays finger printed contig map (http://www.gramene.org/Zea_mays/) on chromosome 5L, contig 234, position 133.32 Mb. A rf2e1 homolog, found by BLASTn analysis (Z. mays PCO083188_ov mRNA, accession AY107915), is mapped on chromosome 5L, contig 240, position 145.42 Mb. The distance between the serk2 and the rf2e1 sequences is 12 Mb.
Large cDNAs:
Approximately 11% of maize genes contain repetitive sequences in their introns (Haberer et al. 2005) and will not be suitable as FISH probes without removal of the repeats. Because fully processed mRNAs do not contain introns and are therefore likely to contain less repetitive DNA, a database search was conducted for large maize cDNA sequences to use as FISH probes. Thirty-six candidate mRNA sequences >4000 bp were identified, including both mapped and unmapped genes. Of these, several with cDNA sequences >6000 bp were selected for further analysis, including Z. mays B73 calpain-like protein (dek1) (7110 bp), the unconventional myosin heavy chain (myo1) (5375 bp), and the acetyl-coenzyme A carboxylase (acc1) (7324 bp) genes. The myo1 gene had not previously been localized and the maize genome contains two highly similar acc genes (Ashton et al. 1994) present on chromosome arms 2L (acc2) and 10L (acc1) (http://www.maizegdb.org). The resulting PCR products were used as FISH probes (Figure 1). Probe dek1 labeled the expected interstitial position on chromosome 1S and additional signal was seen at the NOR due to contaminating cDNA from the rDNA genes (supplemental Figure 2 at http://www.genetics.org/supplemental/). By using RNA that is enriched for poly(A)-containing mRNA as the RT–PCR template, the NOR hybridization signal was eliminated (Figure 1, supplemental Figure 1, E and F, at http://www.genetics.org/supplemental/), and purified mRNA was used as the template for subsequent RT–PCR reactions. The myo1 probe produced a signal on the distal end of chromosome 3L. The acc1/acc2 probe hybridized near the centromere on chromosome 2L and at an interstitial position on chromosome 10L (Figure 1). Thus, the position of the myo1 gene has been determined and the positions of dek1, acc1, and acc2 genes have been detected using FISH.
Pooled PCR products from BACs:
Although extensive BAC libraries exist for maize, the abundance of dispersed repetitive elements prevents the direct use of maize BACs as FISH probes. Many BAC clones have been sequenced as part of the ongoing maize genome sequencing effort, allowing the identification of unique or genic regions using sequence analysis software. Pooling multiple low-copy sequences from a BAC sequence would allow FISH to a genomic target of sufficient size to be readily detectable and free of background signal.
To develop a FISH marker for chromosome 8, the 136.9-kb BAC clone sequence AC157487 was selected. Four unique regions with sizes of 7.5, 13.4, 7.0, and 8.4 kb were identified after RepeatMasker analysis (Figure 2A). Each region was analyzed using the BLASTn program and sequences with homology to plant cDNAs or mRNAs were selected for primer design. These regions were expected to be conserved among maize varieties. Seven PCR products were labeled as FISH probes and individually tested on chromosome spreads. Three PCR products showing no background were combined to produce a probe totaling 8.7 kb in length that readily detected a specific region on chromosome 8L (Figure 1, Figure 2B, Table 1). Four PCR products showed nonspecific hybridization due to elements being present, which can be missed during RepeatMasker analysis. As additional repetitive elements are added to the Repeat Masker library, selection of maize unique sequences for FISH probe development will be more effective. We anticipate that many additional probes will be produced in this fashion and propose the following naming system. Probes will be designated by the chromosome arm and the GenBank accession number used in their design. Thus, a probe produced by pooling the PCR products is named BAC8L–AC157487. Because only one probe on 8L is used in this study, an abbreviated form of the name, BAC8L, will be used.
A similar approach was applied to develop a probe for chromosome 9S using sequence AF448416 from a BAC clone containing the bz1 regions (106.2 kb). RepeatMasker and BLASTn analysis was performed in the same manner as noted above. Because the bz1 region has been well characterized (Fu and Dooner 2002; Brunner et al. 2005), it was possible to select sequences shared among three different inbreds—B73, Mo17, and McC—for FISH probe development. Seven PCR primer pairs were designed. Five PCR products corresponding to regions of genes stk1, stc1, znf, tac7077, and uce2 showed low background as FISH probes and were pooled to produce a readily detectable 12.3-kb probe for the chromosome 9 bz region (Table 2, Figure 1). The other two PCR products were not used because they produced high background. This probe, BAC9S–AF448416, will be referred to as BAC9S in this report.
Karyotyping:
By combining the new probes produced for chromosomes 1S, 5L, 7S, 8L, and 9S with others described previously, including the p1 gene on 1S (Yu et al. 2007), the 5S ribosomal gene cluster on 2L (Kato et al. 2004), the rp3 disease resistance gene cluster on 3L (Kato et al. 2006), the Cent4 repeat cluster near the centromere of chromosome 4 (Kato et al. 2004), the expansin B11 gene cluster expB11 on 5L (Valdivia et al. 2007), the 45S (NOR) ribosomal gene cluster on 6S (Kato et al. 2004), the expansin B9 gene cluster expB9 on 9L (Valdivia et al. 2007), and the rp1 disease resistance gene cluster on 10S (Kato et al. 2006), a collection of single-locus FISH probes that includes at least one on each chromosome has been assembled (Table 2). By combining selected members of this collection, labeled in different colors, it is possible to identify each chromosome in the maize karyotype. The probe cocktail was successfully applied to chromosomes of inbred lines B73, Oh43, and KYS (KYS is shown in Figure 3). The position of each probe on the respective chromosome of inbred B73 was measured and an idiogram was constructed (Figure 4).
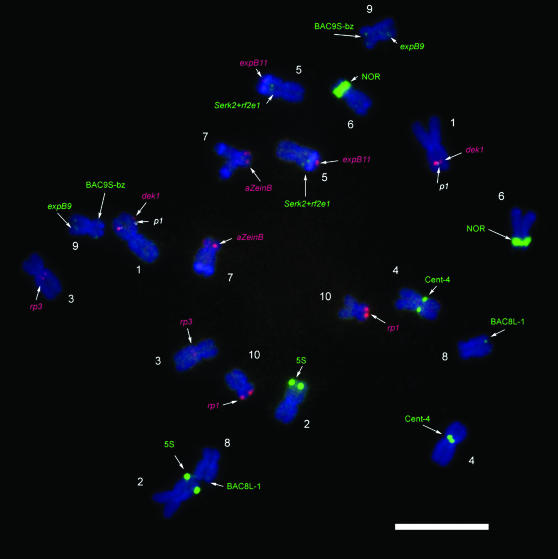
Figure 3.—
Inbred KYS chromosomes identified using small-target probes. FISH with small-target probes including dek1 (1S, red), p1-wr (1S, white), rp3 (3L, red), serk2+rf2e1 (5L, green), expB11 (5L, green), α-zeinB (7S, red), BAC8L (8L, green), BAC9S (9S, green), expB9 (9L, green), and rp1 (10S, red) and with probes to the repetitive elements 5S rDNA (2L, green), 45S rDNA (NOR, 6S, green), and Cent4 (green). See Table 1 for details on each probe. Bar, 10 μm.
Figure 4.—
Idiogram of Z. mays B73 chromosomes showing average relative chromosome lengths (as percentages) and positions of small-target probes on somatic chromosomes from inbred line B73. The asterisk by chromosome 4 indicates the position of a second site of hybridization to a 19-kDa zein gene probe and the asterisk by chromosome 9 indicates a minor site of hybridization to expB10. The colors used to indicate the probe positions correspond to the colors in Figure 3 except probes not used for karyotyping, which are shown in black.
Probe extension to other maize lines and relatives:
Because single-locus FISH probes detected B73 genes or gene clusters whose function is likely conserved, it was expected that they would hybridize in other maize lines and related species. To confirm this supposition, the probes dek1, serk2+rf2e1, BAC8L, and BAC9S were applied to the inbreds KYS and Oh43 and produced signals in the expected locations (supplemental Figure 1 at http://www.genetics.org/supplemental/). Several of the probes in the single-locus collection, including α-zeinA, rp1, and rp3, were previously shown to hybridize to unique locations in Tripsacum and Z. diploperennis (Lamb and Birchler 2006). The remaining probes from the single-locus collection—dek1, serk2+rf2e1, BAC8L, BAC9S, expB11, α-zeinB, acc1/acc2, and myo1—were applied to chromosome spreads from F1 hybrids between maize and wild relatives, including Z. luxurians, Z. diploperennis, and T. dactyloides, and to a “tri-species hybrid” containing chromosomes from Z. mays, Z. diploperennis, and T. dactyloides. The presence of the maize chromosomes in these hybrids provides a positive control for ensuring that the conditions were optimal for signal detection. For all the probes, signal could be detected on the chromosomes from the wild relatives. Several examples are included in Figure 5 and supplemental Figure 2 at http://www.genetics.org/supplemental/. The number of signals in the wild Zea species was the same as that in maize for each probe. In Tripsacum, rp1, rp3, α-zeinA (Lamb and Birchler 2006), α-zeinB, myo1, and acc1/acc2 probes all produced the same number of signals per haploid genome as in maize (Figure 5, supplemental Figure 2 at http://www.genetics.org/supplemental/). The other probes produced more signals per haploid genome than Zea did: BAC8L (three signals), dek1 (two signals), and serk2+rf2e1 (three signals). Because BAC8L is a mixture of three detectable PCR products (3125, 2179, and 3353 bp long), the three sites could indicate three homologous regions or result from separation of the genomic locations relative to their positions in maize.
Figure 5.—
Extension of small-target FISH probes to wild relatives of Z. mays. Small-target probes are indicated by arrowheads. The gray-value images depict the small-target labeling alone. Bars, 10 μm. (A) Maize × Z. diploperennis F1 hybrid chromosomes labeled with BAC8L (red) and (B) ExpB11 (red). The Grande retrotransposon probe (green) hybridizes strongly to maize chromosomes and with intermediate intensity to Z. diploperennis chromosomes. (C) “Tri-species” hybrid containing a haploid set of chromosomes from maize (n = 9, as chromosome 2 is missing), Z. diploperennis (n = 10), and T. dactyloides (n = 18), labeled with myo1 (red) and a Tripsacum-specific retroelement probe, TC#25 (green). Three myo1 signals are observed, two on Zea chromosomes and one on a Tripsacum chromosome. (D) Maize × Z. luxurians F1 hybrid labeled with the serk2+rf2e1 probe (red) and the 180-bp knob probe (green). Knob signals in Z. luxurians are located at the ends of chromosomes whereas maize knob signals are interstitial.
DISCUSSION
Developing FISH probes:
Because current techniques allow routine detection of 3000-bp genomic targets in maize (Kato et al. 2006; Wang et al. 2006; Yu et al. 2007), it is possible to generate FISH probes via PCR amplification of small unique sequences avoiding abundant repetitive elements. Development of probes is limited only by the ability to recognize the unique sequences in the maize genome. In this study, we used various resources to choose targets >3000 bp for which FISH signals will be easily detectable landmarks on maize somatic chromosomes.
To identify unique sequences on specific maize chromosomes, databases were searched for gene sequences that have been mapped to specific chromosomes. The entire gene region, including the introns, was used as a FISH probe. The sequences of physically positioned BACs were analyzed using RepeatMasker and BLASTn, allowing the unique regions, which were conserved among maize lines and other plant species, to be selected and pooled. Long cDNAs were identified using a search algorithm. Coding regions from genes found in tandem clusters were also used. Probes produced from unique sequences are highly specific and showed little or no background hybridization in spite of low stringency of hybridization and washing: 64–67% for probes serk2+rf2e1, BAC8L, and BAC9S (calculated according to formulas and tables from Schwarzacher and Heslop-Harrison 2000).
By selecting the unique regions, any sequence of sufficient size can be used to design FISH probes. This approach should be applicable to any species for which sequence data are available. In particular, it will provide an alternative to BAC–FISH for developing cytological markers in species with large numbers of repetitive elements in their genomes. When extensive cDNA sequences are available, the identification of large cDNAs would provide an effective method for generating a collection of useful FISH probes. Also, PCR amplification of genomic DNA with primers designed from cDNAs <3 kb in length will produce products that include the introns. If these products are unique and >3 kb, they will be suitable as FISH probes.
Numerous sequences of unmapped maize genomic DNAs, mRNAs, and cDNAs are currently available. The technique described here could be applied to determine their cytological position. Other probes may be included in different colors to identify each chromosome in addition to the sequence of unknown position. For the common inbred lines B73, Oh43, and KYS, all chromosomes can be identified with the inclusion of the CentC and TAG microsatellite probes in one channel and DAPI counterstaining. As an alternative, a probe collection consisting of 5S, rp3, Cent4, expB11, BAC8L, expB9, and rp1, all labeled in one color, will allow each chromosome to be identified on the basis of signal intensity and position as well as chromosome size. Either approach uses only two fluorescent channels for karyotyping, leaving others free for experimental probes. The utility of such an approach has been demonstrated by determining the chromosomal location of the myo1 gene in this work and numerous transgene insertions in previous work (Yu et al. 2006). This approach will be particularly useful for genetic loci that are refractory to traditional mapping, including genes near centromeres where recombination rates are low (Anderson et al. 2003). FISH could also help confirm the predicted contig alignments of physical maps.
As more maize sequences are determined, it will be possible to use them to design additional FISH probes. We have demonstrated that several separated unique sequence regions from a chromosomal region that are individually too small to be readily detected can be pooled to create a detectable FISH probe. However, current identification of unique sequences using the RepeatMasker software relies on comparisons to repetitive element libraries. Because the current library is incomplete, candidate probes selected from genomic sequence may still produce high background signal. Therefore, the utility of each candidate probe needs to be verified before combining sequences. Software that combines identification of repetitive elements and selection of primers for FISH probe synthesis from genomic regions of the human genome has been written (Navin et al. 2006). This software does not rely on previously characterized repeat libraries. Instead, it assigns sequences a repetitiveness score on the basis of the number of times they are present in a sequence database (Navin et al. 2006). Similar software could aid in the development of additional probes for maize.
Large amounts of sequence are not available for all species of scientific or agronomic interest. One example is Tripsacum, which is a valuable forage crop. Previous work demonstrated that retrotransposons could be used instead of genomic in situ hybridization to distinguish chromosomes from the two genera in hybrids (Lamb and Birchler 2006). Also, repetitive satellite sequences from maize were used to label heterochromatic regions, including centromeres and subtelomeric regions in Tripsacum (Lamb and Birchler 2006). Little Tripsacum sequence is available to design single-locus probes, but maize genes produce FISH signals on Tripsacum chromosomes. In some cases the number of signals per haploid Tripsacum genome was the same as that for maize but for others it was greater. The additional loci could indicate that the Tripsacum genome is a tetraploid compared to maize or could reflect individual gene duplication events. As more FISH probes are developed in maize, it will be possible to conduct a thorough cytological comparison of the two species. This study emphasizes the potential of using sequence information from one species to develop cytological tools for relatives and could be applied to other groups with one member that has been well characterized genomically.
Karyotyping using single-locus probes:
Chromosome identification is a valuable aspect of any cytogenetic investigation. Karyotype development in most plant species has usually relied on the arm-length ratios, chromosome size, and FISH probes that detect repetitive elements such as ribosomal repeats and chromosome-specific heterochromatic elements. However, differences in the presence and abundance of heterochromatic elements can significantly alter the total length of chromosomes as well as the arm-length ratios among different maize lines (Kato et al. 2004). Therefore, single-locus karyotyping systems that use invariant sequences are more reliable.
In maize, extraordinary sequence variation exists among different lines (Song and Messing 2003; Brunner et al. 2005). Much of this variation is attributable to large transposable elements, known as Helitrons, which capture and move gene fragments (Gupta et al. 2005; Lai et al. 2005; Morgante et al. 2005). As a result, low-copy sequences are present in different genetic locations among maize varieties (Morgante et al. 2005). FISH provides a powerful tool for evaluating the variation in sequence position among lines or species. However, among homologous regions of different maize lines, genes that are collinear with the orthologous rice region were present in all varieties examined (Lai et al. 2005). This finding suggests that much of the polymorphism for maize gene position involves nonfunctional genes or fragments. None of the sequences used in this study produced signals at variable locations in the maize lines examined.
We have developed a collection of FISH probes that detect specific genetic loci on maize somatic chromosomes and that are conserved among different maize lines and some related species. Combinations of probes that allow karyotype assignment in maize and in other Zea species can be assembled. Additionally, individual probes can be used in Tripsacum as a part of a karyotyping system for that genus in the future. The ability to identify chromosomes in any maize line will allow researchers to determine the location of genes, transgenes, unanchored BACs, new transposon insertions, and other genomic features that are cytologically detectable. Because the probes are generally based on genes that are conserved, it is likely that many maize single-locus probes could be applied to even more distantly related species. Including additional probes will lead to the development of a high-density cytogenetic map allowing large genomic rearrangements to be visualized in maize lines and among maize relatives. Creating pools of adjacent sequences could be used to develop chromosome banding or whole-chromosome paints for maize. In conclusion, analyzing genomic sequence to develop FISH probes is a powerful approach to cytological investigation that can be extended to other species.
Acknowledgments
We thank Ed Coe, Georgia Davis, Doug Davis, Karen Cone, and Mary Schaeffer (University of Missouri) for their help with maize physical maps. We thank Patrice Albert for assistance. This work was supported by a grant (DBI 0423898) from the National Science Foundation Plant Genome Initiative.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, L. K., G. G. Doyle, B. Brigham, J. Carter, K. D. Hooker et al., 2003. High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton, A. R., C. L. Jenkins and P. R. Whitfeld, 1994. Molecular cloning of two different cDNAs for maize acetyl CoA carboxylase. Plant Mol. Biol. 24: 35–49. [DOI] [PubMed] [Google Scholar]
- Bauer, M. J., and J. A. Birchler, 2006. Organization of endoreduplicated chromosomes in the endosperm of Zea mays L. Chromosoma 115: 383–394. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and E. A. Kellogg, 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., K. Fengler, M. Morgante, S. Tingey and A. Rafalski, 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., A. Gallavotti, G. A. Stryker, R. J. Schmidt and S. K. Lal, 2005. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol. Biol. 57: 115–127. [DOI] [PubMed] [Google Scholar]
- Haberer, G., S. Young, A. K. Bharti, H. Gundlach, C. Raymond et al., 2005. Structure and architecture of the maize genome. Plant Physiol. 139: 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., B. S. Gill, G. L. Wang, P. C. Ronald and D. C. Ward, 1995. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92: 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., J. C. Lamb and J. A. Birchler, 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., P. S. Albert, J. M. Vega and J. A. Birchler, 2006. Sensitive FISH signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech. Histochem. 81: 71–78. [DOI] [PubMed] [Google Scholar]
- Kim, J. S., K. L. Childs, M. N. Islam-Faridi, M. A. Menz, R. R. Klein et al., 2002. Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45: 402–412. [DOI] [PubMed] [Google Scholar]
- Kim, J. S., M. N. Islam-Faridi, P. E. Klein, D. M. Stelly, H. J. Price et al., 2005. Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171: 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., P. Fransz and H. De Jong, 2003. Cytogenetic tools for Arabidopsis thaliana. Chromosome Res. 11: 183–194. [DOI] [PubMed] [Google Scholar]
- Koumbaris, G. L., and H. W. Bass, 2003. A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J. 35: 647–659. [DOI] [PubMed] [Google Scholar]
- Lai, J., Y. Li, J. Messing and H. K. Dooner, 2005. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102: 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J. C., and J. A. Birchler, 2006. Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics 173: 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J. C., A. Kato, W. Yu, F. Han, P. S. Albert et al., 2006. Cytogenetics and chromosome analytical techniques, pp. 244–248 in Floriculture, Ornamental and Plant Biotechnology, edited by J. A. Teixeira da Silva. Global Science Books, London.
- Lamb, J. C., J. M. Meyer, B. Corcoran, A. Kato, F. Han et al., 2007. Distinct chromosomal distributions of highly repetitive sequences in maize. Chromosome Res. 15: 33–49. [DOI] [PubMed] [Google Scholar]
- Li, L., and K. Arumuganathan, 2001. Physical mapping of 45S and 5S rDNA on maize metaphase and sorted chromosomes by FISH. Hereditas 134: 141–145. [DOI] [PubMed] [Google Scholar]
- Morgante, M., S. Brunner, G. Pea, K. Fengler, A. Zuccolo et al., 2005. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37: 997–1002. [DOI] [PubMed] [Google Scholar]
- Navin, N., V. Grubor, J. Hicks, E. Leibu, E. Thomas et al., 2006. PROBER: oligonucleotide FISH probe design software. Bioinformatics 22: 2437–2438. [DOI] [PubMed] [Google Scholar]
- Reeves, A., and J. Tear, 2000. MicroMeasure for Windows, version 3.3 (http://www.colostate.edu/depts/biology/micromeasure).
- Rivin, C. J., C. A. Cullis and V. Walbot, 1986. Evaluating quantitative variation in the genome of Zea mays. Genetics 113: 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadder, M. T., and G. Weber, 2001. Karyotype of maize (Zea mays L.) mitotic metaphase chromosomes as revealed by fluorescent in situ hybridization (FISH) with cytogenetic DNA markers. Plant Mol. Biol. Rep. 19: 117–123. [Google Scholar]
- Sadder, T., and G. Weber, 2002. Comparison between genetic and physical maps in Zea mays L. of molecular markers linked to resistance against Diatraea spp. Theor. Appl. Genet. 104: 908–915. [DOI] [PubMed] [Google Scholar]
- Sadder, M. T., N. Ponelies, U. Born and G. Weber, 2000. Physical localization of single-copy sequences on pachytene chromosomes in maize (Zea mays L.) by chromosome in situ suppression hybridization. Genome 43: 1081–1083. [PubMed] [Google Scholar]
- Schwarzacher, T., and P. Heslop-Harrison, 2000. Practical in Situ Hybridization. BIOS Scientific Publishers, Oxford.
- Smit, A. F., R. Hubley and P. Green, 2004. RepeatMasker Open-3.0 (http://www.repeatmasker.org).
- Smith, S. M., A. J. Pryor and S. H. Hulbert, 2004. Allelic and haplotypic diversity at the rp1 rust resistance locus of maize. Genetics 167: 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2002. Contiguous genomic DNA sequence comprising the 19kD zein gene family from maize. Plant Physiol. 130: 1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia, E. R., J. Sampedro, J. C. Lamb, S. Chopra and D. J. Cosgrove, 2007. Recent proliferation and translocation of pollen group 1 allergen genes in the maize genome. Plant Physiol. (in press). [DOI] [PMC free article] [PubMed]
- Wang, C. J., L. Harper and W. Z. Cande, 2006. High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18: 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, C. A., T. E. Richter, N. C. Collins, M. Nicolas, H. N. Trick et al., 2002. Genetic and molecular characterization of the maize rp3 rust resistance locus. Genetics 162: 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, Y. M., D. W. Hu, B. A. Larkins and R. Jung, 2001. Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., R. B. Meeley and D. J. Cosgrove, 2001. Analysis and expression of the alpha-expansin and beta-expansin gene families in maize. Plant Physiol. 126: 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., J. C. Lamb, F. Han and J. A. Birchler, 2006. Telomere-associated chromosomal truncation in maize. Proc. Natl. Acad. Sci. USA 103: 17331–17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., J. C. Lamb, F. Han and J. A. Birchler, 2007. Cytological visualization of transposable elements and their transposition pattern in somatic cells of maize. Genetics 175: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, M. S., M. N. Islam-Faridi, D. G. Czeschin, Jr., R. A. Wing, G. E. Hart et al., 1998. Physical mapping of the liguleless linkage group in Sorghum bicolor using rice RFLP-selected sorghum BACs. Genetics 148: 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]