Abstract
Glutathione (GSH), l-γ-glutamyl-l-cysteinyl-glycine, is the major low-molecular-weight thiol compound present in almost all eukaryotic cells. GSH degradation proceeds through the γ-glutamyl cycle that is initiated, in all organisms, by the action of γ-glutamyl transpeptidase. A novel pathway for the degradation of GSH that requires the participation of three previously uncharacterized genes is described in the yeast Saccharomyces cerevisiae. These genes have been named DUG1 (YFR044c), DUG2 (YBR281c), and DUG3 (YNL191w) (defective in utilization of glutathione). Although dipeptides and tripeptides with a normal peptide bond such as cys-gly or glu-cys-gly required the presence of only a functional DUG1 gene that encoded a protein belonging to the M20A metallohydrolase family, the presence of an unusual peptide bond such as in the dipeptide, γ-glu-cys, or in GSH, required the participation of the DUG2 and DUG3 gene products as well. The DUG2 gene encodes a protein with a peptidase domain and a large WD40 repeat region, while the DUG3 gene encoded a protein with a glutamine amidotransferase domain. The Dug1p, Dug2p, and Dug3p proteins were found to form a degradosomal complex through Dug1p–Dug2p and Dug2p–Dug3p interactions. A model is proposed for the functioning of the Dug1p/Dug2p/Dug3p proteins as a specific GSH degradosomal complex.
GLUTATHIONE (GSH), l-γ-glutamyl-l-cysteinyl-glycine, is the major low-molecular-weight thiol compound present in almost all eukaryotic cells (Meister and Anderson 1983; Fahey and Sundquist 1991) at intracellular concentrations ranging from 0.1 to 10 mm (Hwang et al. 1992). This is in contrast to other redox couples that are in micromolar concentrations in the cell (Holmgren et al. 1978). GSH thus acts as the principal redox buffer, plays an important role in oxidative stress response and in the detoxification of metals and xenobiotics, and influences—through redox—several essential processes such as gene expression, cell proliferation, and apoptosis (Penninckx and Elskens 1993; Arrigo 1999; Fang et al. 2002). The two important chemical properties from which glutathione derives its importance in the cell are its low redox potential (Ostergaard et al. 2004) and the stability of the tripeptide provided by the unusual γ-glutamyl bond, making it resistant to peptidases in the cell and allowing it to exist at high concentrations in the cell (Ganguly et al. 2003).
GSH levels (and the ratio of the oxidized and reduced forms of glutathione) need to be carefully maintained in the cell. In addition to its biosynthesis, degradation, and consumption in different processes, glutathione levels are altered by its compartmentalization and efflux from the cell (Perrone et al. 2005). In addition to biosynthesis of GSH, which occurs in the cytoplasm through the sequential action of two cytosolic enzymes, γ-glutamyl cysteine synthase and glutathione synthase (Meister and Anderson 1983), GSH can also be transported from the extracellular medium through specific transporters (Bourbouloux et al. 2000). These multiple processes combine to maintain glutathione homeostasis in the cell. GSH deficiency in the cell has been associated with many disease states that include liver diseases, macular eye degeneration, Alzheimer's, aging, and HIV infections (Wu et al. 2004). Higher levels of glutathione have also been shown to lead to glutathione toxicity at least in yeast (Srikanth et al. 2005).
In eukaryotes, glutathione is essential for growth. In the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, deletion of the first enzyme in GSH biosynthesis leads to growth stasis (unless supplied with exogenous glutathione) (Wu and Moye-Rowley 1994; Grant et al. 1996; Chaudhuri et al. 1997), while in mice deletion of the first enzyme leads to embryonic lethality (Shi et al. 2000). Overproduction of the rate-limiting enzyme in glutathione biosynthesis has recently been demonstrated as leading to increased longevity in Drosophila melanogaster (Orr et al. 2005).
GSH biosynthesis and metabolism proceeds through the γ-glutamyl cycle proposed by Orlowski and Meister (1970). γ-Glutamyl transpeptidase (γ-GT) catalyzes the first step in the degradation of glutathione in this cycle, which involves the cleavage and transfer of the γ-glutamyl moiety from glutathione to an acceptor amino acid (or hydrolysis to release glutamate) and release of cysteinyl-glycine. Ever since the γ-glutamyl cycle was proposed, the degradation of glutathione has been thought to be initated by the action of γ-glutamyl transpeptidase. The possibility of an alternative enzyme (or pathway) for glutathione degradation has never been carefully examined.
We have recently provided genetic evidence for the existence of an alternative pathway for GSH degradation independent of γ-GT (Kumar et al. 2003b). This was demonstrated through the use of cells disrupted in the ECM38 gene encoding the γ-GT enzyme. S. cerevisiae encodes a single enzyme for γ-GT (encoded by the ORF ECM38/CIS2/YLL299w). These cells retained the ability to utilize glutathione as a sulfur source, demonstrating that an alternate pathway for GSH degradation exists in yeast cells. This alternative pathway for GSH degradation allowed the yeast cell to utilize GSH efficiently as a sole source of sulfur (Kumar et al. 2003b). However, while providing new insights into the physiology of GSH metabolism, this finding has also raised several questions regarding the pathway and the exact peptidases that might be mediating this turnover. Since under normal growth conditions GSH is turned over minimally (Mehdi and Penninckx 1997; Kumar et al. 2003a), the nature of this pathway and its components is intriguing, especially since GSH is the principal redox buffer and performs numerous functions that are essential for the viability of living cells. The identification of the enzymes mediating GSH turnover in yeast through the alternative pathway is therefore an important key to understanding the exact physiology of GSH metabolism and homeostasis in cells. In this study we have attempted to investigate these questions and to identify the participants in this novel pathway affecting this very important cellular metabolite.
MATERIALS AND METHODS
Chemicals and reagents:
Glutathione and γ-glutamylcysteine were purchased from Sigma-Aldrich. Cysteinylglycine was purchased from Bachem and α-glutamylcysteinylglycine was custom synthesized by Gensescript. Restriction enzymes and Vent DNA polymerase were from New England Biolabs (Beverly, MA). Oligonucleotides were purchased from BioBasic (Canada). The antibodies used in this study were the following: anti-HA (6E2) mouse mAb (no. 2367, cell signaling), anti-Myc (9B11) mouse mAb (no. 2276, cell signaling), anti-His (27E8) mouse mAb (antibodies against the 6xHis epitope) (no. 2366, cell signaling), and HRP linked anti-mouse immunoglobulin G (IgG) (no. 7076, cell signaling).
Yeast strains, constructions, and growth conditions:
The yeast strains used in this study and their sources are shown in supplemental Table 1 at http://www.genetics.org/supplemental/. Yeast cells were grown at 30° in YPD medium, synthetic defined medium (SD), or complete medium (CM) (Rose et al. 1990). Glutathione, methionine, and cysteine or other peptides were used at a concentration of 200 μm. The yeast strains constructed during this study are described below:
ABC1388 (met15Δlap1lap2lap3lap4) was created by disrupting the MET15 gene in the ABC1339 (lap1lap2lap3lap4) strain by PCR-mediated gene disruption using the KanMX2 module (Wach et al. 1994). The KanMX2 cassette was amplified from plasmid ABE333 (pFA6-KanMX2) using primers MET15-DEL1 and MET15-DEL2. ABC1390 (met15Δpre3T20A) was similarly created by disrupting MET15 in ABC1356 (pre3T20A), and ABC1391 (met15Δpre4-1) was created by disrupting MET15 in the ABC1357 (pre4-1) strain. The disruptions were confirmed by checking the lack of growth on SD plates without any organic sulfur source.
ABC1720 (met15Δcps1ΔkexΔ yb139wΔprc1Δ) lacking four carboxypeptidases was created by sequential disruption of KEX1, YBR139w, and PRC1 genes in a cps1Δmet15Δ strain (ABC1281). The KEX1 gene was disrupted using the URA3 marker. The URA3 cassette was amplified from plasmid ABE150 (pSP2) by using primers KEX1-DEL1 and KEX1-DEL2. Disruptions were confirmed by PCR using primers KEX1CF-FOR and KEX1CF-REV. The YBR139w gene was disrupted using the LEU2 marker. LEU2 cassette was amplified from plasmid ABE149 (pSP1) by using primers YBR139w-DEL1 and YBR139w-DEL2. Disruptions were confirmed by PCR using primers YBR139wCF-FOR and YBR139wCF-REV. The PRC1 gene was disrupted by using a prc1Δ∷HIS3 disruption plasmid (ABE1660). The plasmid ABE1660 was digested with BamHI and transformed in ABC1719 (met15Δ cps1Δ kex1Δ ybr139wΔ). The disruptions were confirmed by checking for lack of carboxypeptidase (CpY) activity by performing the N-acetyl-dl-phenylalanine β-naphthyl ester overlay test (Jones 2002).
Mutant isolation, genetic complementation studies, and yeast DNA isolation:
Ethyl methanesulfonate (EMS) mutagenesis was performed to isolate yeast mutants defective in utilizing GSH as a sole source of sulfur. ABC1083 (met15Δecm38Δ) was taken as the parent strain to isolate mutants that failed to grow on SD plates containing GSH as the sole source of organic sulfur but which could grow well when supplied with methionine or cysteine. The ecm38Δ background was chosen to eliminate any residual contribution to degradation from the γ-glutamyltranspeptidase enzyme. The EMS mutagenesis was done according to standard protocol (Lawrence 2002) and 90% killing was achieved following incubation of wild-type cells in 1.5% EMS for 45 min at 30°. The EMS mutagenized culture was plated onto ∼50 YPD plates, which were replica plated to media containing GSH as the sole sulfur source; mutant clones were purified by single-colony purification and patched onto selective media plates to confirm mutants with the desired phenotype.
Mutants specifically defective in utilization of GSH were backcrossed with a wild-type parent of the opposite mating type (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), and complementation analyses of resulting backcrossed spores were carried on by diploid formation. Complementation analysis was also carried on with a met15Δhgt1Δ mutant. The genes corresponding to the mutants were isolated from a yeast genomic library by functional complementation phenotypic with GSH as the sole source of organic sulfur.
Yeast genomic DNA and plasmid DNA isolation were carried out by the glass-bead lysis method (Kaiser et al. 1994) and yeast transformations were carried out by the lithium acetate method (Ito et al. 1983).
Glutathione toxicity assay:
Glutathione toxicity assay was carried out by the spot dilution assay. The met15Δ, met15Δdug3Δ, met15Δecm38Δ, and met15Δecm38Δdug3Δ strains were transformed with the plasmid pTEF-HGT1 and control vectors. The transformants were grown in minimal medium and dilution spotting was carried out on minimal media plates containing glutamate as a nitrogen source and methionine (200 μm) and glutathione across a range of concentrations.
Cloning of genes and their manipulation:
The DUG1, DUG2, and DUG3 ORFs, the yeast two-hybrid fusions, the promoter-LacZ fusions, and the tagging constructs (as listed in supplemental Table 2 at http://www.genetics.org/supplemental/) were created first by PCR amplifying the relevant regions using the primers listed in supplemental Table 3 (at http://www.genetics.org/supplemental/) and digestion by the appropriate restriction enzymes, followed by cloning into the appropriate vectors using standard protocols. Sequencing was done using gene-specific oligos (data not shown) on an ABI Prism 310 sequencer per manufacturer's protocol.
Detection of thiol intermediates of GSH degradation in dug mutant extracts:
Wild-type and dug1Δ, dug2Δ, and dug3Δ strains were grown in SD media supplemented with 0.2 mm glutathione at 30° for 16 hr, chilled in ice for 30 min, and washed in distilled water. The cell pellet was resuspended in 5% sulfosalicylic acid, and the cell extract was prepared using glass-bead lysis method, and adjusted to pH 7.0 using 10 n NaOH. An equal amount of the cell extracts was labeled with thiol-specific fluorescent labeling reagent monobromobimane in TE buffer (pH 8.0) following the protocol described earlier (Fahey and Newton 1987). The labeled sample was diluted in mobile phase and injected for analysis in a Shimadzu (Tokyo) SCL-10Avp HPLC system with a reverse phase C18 column (150 × 4.6 mm). Buffer A consisted of 0.25% glacial acetic acid in water, pH ∼3.5, and 0.22 μm filtered. Buffer B (B) was HPLC grade methanol. With a flow rate of 0.6 ml/min at 25°, the linear gradients were 15–23% B, 0–15 min; 23–42% B, 15–45 min; 42–75%, 45–55 min; 75–15%, 55–65 min; and 15%, 85 min. The fluorescence was detected using a RF-10Axl detector with excitation at 360 nm and emission at 490 nm. The data were acquired and analyzed using Shimadzu CLASS VP software. Cysteine, γ-gly-cys, and GSH showed 14-, 23.5-, and 25-min retention times, respectively. Cys and cys-gly peaks were detected at the same positions under these conditions.
Yeast two-hybrid analyses:
The DupLex-A yeast two-hybrid system (OriGene Technologies, MD) based on the system developed by R. Brent and co-workers (Gyuris et al. 1993) was used in this study. In this system, the “bait plasmid” is pEG202, in which the test protein is fused to the LexA DNA-binding domain. The “prey plasmid” is pJG4.5, in which the Gal4p protein activation domain is fused to the second test protein. Interaction of the test proteins with bait/prey plasmids leads to the formation of a functional transcription activator. For the reporter, two systems are used: (1) growth on the −Leu plate and (2) color development on the +X-Gal plate. For growth on −Leu, the strain EGY48, transformed with pEG202 (pBait), pJG4.5 (pPrey), and pSH18-34, which contained 6LexA operators upstream of the LEU gene, was used. In X-Gal color development, pEG202 (pBait), pJG4.5 (pPrey), and pSH18-34 plasmids were transformed in the EGY48 strain. This pSH18-34 plasmid contained 8LexA operators upstream of LacZ gene. The EGY48 yeast strain, transformed with a specific set of three yeast two-hybrid clones, was selected on CM + Glc with appropriate selection. The transformants were patched on CM − Leu and CM + X-gal assay plate with appropriate selection. The growth or color development was monitored by incubating the plates at 30° for an optimal number of days. The auto-activation potential of each bait construct (pEG202-X) was examined by analyzing the ability of the transformant to grow on CM − Leu plate with appropriate selection (LEU2 activation) and by using a lacZ reporter vector, pSH18-34 (X-gal color assay) as well.
Immunoprecipitation assays:
The yeast strain ABC710, a protease-deficient strain (deficient in protease A, protease B, and protease C), was used as a host strain in these studies. Yeast transformants bearing multiple plasmids, expressing the tagged proteins, were grown in SD media with appropriate selection up to OD600 0.4–0.5 and then harvested at 4°. Cells were washed once with ice-cold yeast protein extraction buffer (20 mm TrisCl (pH 7.5), 20 mm NaCl, 1 mm EDTA, 10% glycerol, 0.2% NP-40) and were resuspended in the same ice-cold yeast protein extraction buffer. Cells were broken using glass-bead lysis method following the protocol as described (Brown et al. 2000). The supernatant was collected after centrifugation at 13,000 rpm for 10 min at 4°. The concentration of the total protein in samples was measured using a Bradford reagent with BSA as the standard. The samples (∼200 μg) were incubated with either anti-HIS mouse mAb or anti-cMyc mouse mAb for ∼2 hr at 4° on an Eppendorf thermo-mixer. The sample was incubated with 2× vol of ProteinG sepharose slurry for ∼1 hr at 4° on an Eppendorf thermo-mixer. Beads were centrifuged and washed three times with phosphate buffer. Samples (beads) were boiled in 5X× SDS–PAGE sample buffer and run on SDS–PAGE (Sambrook et al. 1989) and proteins were transferred to nitrocellulose membrane (Towbin et al. 1979) and immunodetected using primary anti-His mouse mAb, anti-cMyc mouse mAb, or anti-HA mouse mAb (dilution 1:2500) and secondary HRP-conjugated Horse anti-mouse IgG (dilution 1:7500) following the procedure described by Blake et al. (1984) and visualized using an enhanced chemiluminescence kit.
Fluorescence microscopy:
Fluorescence microscopy was performed on an inverted LSM510 META laser scanning confocal microscope (Carl Zeiss) fitted with plan-Apochromat ×100 (numerical aperture, 1.4) oil immersion objective. All measurements were performed with living nonfixed cells. For detection of the Dug1GFP, Dug2GFP, and Dug3GFP fusion proteins, the 488-nm line of an argon ion laser was directed over an HFT UV/488 Beam splitter, and fluorescence was detected using an NFT 490 beam splitter in combination with a BP 505- to 530-band pass filter (Aggarwal and Mondal 2006). Strains expressing various DUGXGFP were grown in SD media, supplemented with GSH and other supplements, to logarithmic phase (OD600 ∼0.5–0.6) and observed under microscope. Images obtained were processed using Adobe Photoshop Version 5.5.
RESULTS
GSH utilization by the alternative pathway requires a functional cysteine utilization pathway, but does not involve known peptidolytic activities:
To obtain insights into the possible components of the alternative pathway, we examined if GSH degradation and utilization as a sulfur source proceeded through the utilization of cysteine, since cysteine was one of the component amino acids of glutathione and appeared to be the most likely route for GSH utilization. Yeast str2Δ strains, deleted for the STR2 gene that encodes cystathionine γ-synthase, cannot utilize cysteine as the sole source of sulfur as it is the first step in the utilization of cysteine (Hansen and Johannesen 2000). The str2Δ strain, in addition to being unable to grow on cysteine as a sole source of sulfur, was unable to utilize GSH as a sole source of sulfur, even in the presence of a functional γ-glutamyl-transpeptidase (data not shown). The inability of the str2Δ strain to grow on either cysteine or GSH suggests that GSH utilization by the alternative pathway requires a functional cysteine utilization pathway and is most likely to proceed through cysteine as an intermediate.
S. cerevisiae has several aminopeptidases, carboxypeptidases, as well as dipeptidases, making them potential candidate peptidases involved in degradation of the tripeptide glutathione in the absence of γ-glutamyltranspeptidase. As cysteine was indicated as the possible intermediate in this pathway, the possibility that this might be released through the action of one of the large number of aminopeptidases or carboxypeptidases in the yeast was highly likely.
We examined the possible role of the major peptidases that has been described in S. cerevisiae in GSH turnover by evaluating strains deleted for the individual genes in the met15Δ background for growth on GSH as a sole source of sulfur. The met15Δ background was chosen since these strains are organic sulfur auxotrophs and cannot use inorganic sulfate as a source of sulfur. They can use cysteine, methionine, or glutathione as sulfur sources. To use GSH as a sulfur source, GSH needs to be degraded into its constituent amino acids. The different deletion strains examined are shown in Table 1. In addition, to eliminate redundancy in function among the aminopeptidases, as well as among the carboxypeptidases, we created strains multiply deficient in these activities as shown in the Table 1. However, no effect on glutathione utilization was detected in any of these strains.
TABLE 1.
Growth of different peptidase disruptants on GSH and methionine
| Type of disruptants | Growth phenotype
|
|||
|---|---|---|---|---|
| Pathway | Gene disrupted/mutated | +MET | +GSH | |
| Vacuolar biogenesis/proteolysis | Single | pep5Δ and pep4Δ | ++ | ++ |
| Vacoular autophagy | Single | apg1Δ, tor1Δ, aut4Δ, aut2Δ, atg15 and apg14 | ++ | ++ |
| Aminopeptidase | Single | lap1Δ, lap3Δ, lap4Δ, aap1Δ, ape3Δ, dap2Δ, and ynl045w | ++ | ++ |
| Multiple | lap1lap2lap3lap4 | ++ | ++ | |
| Carboxypeptidase | Single | ybr139wΔ, prc1Δ cps1Δ, and kex1Δ | ++ | ++ |
| Multiple | ybr139wΔprc1Δcps1Δkex1Δ | ++ | ++ | |
| Proteasomal peptidases | Single | pre3T20A and pre4-1 | ++ | ++ |
Peptidase genes disrupted in met15Δ background were examined for their involvement in utilization of GSH as sole source of sulphur.
The possibility of a role for the vacuole and vacuolar autophagy in utilization of GSH as an exogenous source of sulfur was also evaluated as the vacuoles have a large number of peptidases and vacuolar autophagy plays an important role in the starvation condition for meeting amino acid requirements. However, none of the mutants displayed a selective growth defect on glutathione utilization.
Since the known aminopeptidases, carboxypeptidases, and dipeptidases or vacuolar autophagy were not involved in the utilization of GSH as a sole source of sulfur in S. cerevisiae, we evaluated the possible contribution of the proteasome, the major proteolytic and peptidolytic enzyme present in the cytoplasm that is responsible for the bulk degradation of proteins and peptides for its role in GSH degradation. One of the major activities of the yeast proteasome is peptidylglutamyl–peptide-hydrolyzing (PGPH) activity, which involves cleavage of bonds on the carboxyl side of acidic amino acids in the proteins or peptides and is dependent on two of its subunits, PRE3 and PRE4. Each of these subunits is essential for cell viability (Heinemeyer et al. 1991, 1993; Hilt et al. 1993).
Mutant alleles for PRE3 and PRE4 subunits that lack PGPH activity (Hilt et al. 1993; Heinemeyer et al. 1997) were procured and we created MET15 disruptions in the two mutant strains as well as in the corresponding wild-type strain. Both met15Δ pre3T20A and met15Δpre4-1 strains were found to grow well on GSH just as the wild-type strain and the growth on GSH were also similar to the growth on methionine, suggesting that the peptidylglutamyl–peptide-hydrolyzing activity of the yeast proteasome is not needed for the utilization of GSH as a sulfur source.
Isolation of mutants defective in utilization of glutathione (dug) and the placement of the dug mutants into four complementation groups:
The absence of an apparent involvement of the known peptidases and the known proteolytic pathways indicated either a redundancy of the different pathways or the possibility of a new pathway for glutathione degradation. The possibility also existed that there might be a small redundancy with the known enzyme involved in glutathione degradation (encoded by ECM38), which was masking the detection of the deletion strains that were examined. To address these issues, we decided to attempt the isolation of mutants defective in glutathione degradation.
A met15Δ ecm38Δ (ABC1083) strain was taken as the parent strain to isolate mutants that failed to grow on SD plates having GSH as the sole source of organic sulfur but grew well when supplied with methionine or cysteine in place of GSH. In addition to met15Δ, an ecm38Δ background was chosen to rule out any participation of γ-GT in this process. We carried out the EMS mutagenesis at 90% killing (described in detail in materials and methods), followed by plating on YPD plates and incubation at 30° for 2–3 days. The colonies obtained were replica plated on minimal medium containing glutathione, methionine, or cysteine. Colonies that grew on methionine and cysteine but not on GSH plates were picked up as putative mutants and single colony purified. From among ∼40,000 colonies that were screened, we obtained 45 mutants that failed to grow on SD plates having GSH as the sole source of sulfur but grew well on plates having methionine or cysteine. These 45 mutants were subsequently restreaked for single colonies on plates having GSH or methionine or cysteine. Of the initial 45 isolates, 12 recovered the ability to grow on GSH upon subsequent streaking and these were excluded from further analysis. A total of 33 stable mutants failed to grow on SD plates having GSH as the sole source of sulfur, but grew well on other media.
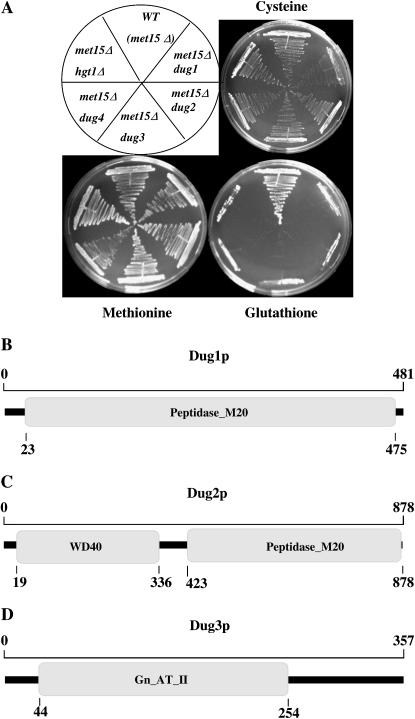
To examine whether the 33 mutants fell into single or multiple complementation groups, complementation analysis was carried out. As the initial mutant isolation was carried out in the MATa mating type, it was necessary to generate the mutants in the opposite MATα background. The mutants were backcrossed with the wild type and mutants in opposite mating types were also obtained. The mutants were then placed into complementation groups by creating diploids and checking the ability of diploids to grow on GSH as a sole source of sulfur. A total of four complementation groups were identified in which all mutants could be placed. The complementation groups were named dug1, dug2, dug3, and dug4 (defective in utilization of glutathione). A total of 23 mutants were in the dug4 group, 5 mutants were in the dug3 group, 3 mutants were in the dug2 group, and 2 mutants were in the dug1 group. The growth of a representative dug mutant strain from each complementation group was checked on a plate containing different organic sulfur sources (Figure 1A).
Figure 1.—
(A) Growth of dug1, dug2, dug3, and dug4 mutant strains along with wild-type (WT) and hgt1Δ strains (in a met15Δ background) on SD plates containing cysteine, glutathione, or methionine at a concentration of 0.2 mm. Schematic of the conserved domains of the three Dug proteins analyzed using NCBI-BLASTp (Altschul et al. 1990) and NCBI-CDD (Marchler-Bauer and Bryant 2004). (B) Dug1p shows homology to peptidase of class M20. (C) Dug2p consists of two distinct domains: an N-terminal WD40 repeat domain and a C-terminal peptidase of class M20. (D) The N terminus of Dug3p shows homology to glutamine amidotransferase, class II (Gln-AT-II) domain. The numbers refers to amino acid residue numbers.
The dug4 complementation group and the glutathione transporter class (hgt1) fall into the same complementation group:
Previous observations in our lab (Bourbouloux et al. 2000) have revealed that deletions in the yeast glutathione transporter gene (HGT1) in a met15Δ background fail to grow on minimal plates having GSH as the sole organic sulfur source, while they grow well on methionine and cysteine, owing to the inability of these mutants to transport GSH. Hence, we expected that one of the complementation groups would carry mutations at the HGT1 gene locus, which encodes the glutathione transporter. One mutant from each of the four complementation groups was transformed with a plasmid having HGT1 (pNP-HGT1) under its native promoter. The transformants were initially selected on plates having methionine and later checked by streaking onto plates having GSH. The ability to grow on GSH was restored in one of the mutants belonging to complementation group dug4. To further confirm this observation, all the mutants were transformed with the HGT1 plasmid. In addition, these results were confirmed with genetic complementation tests. The HGT1 plasmid complemented the growth defect on GSH plates in 23 of 33 total mutants belonging to the dug4 complementation group. Thus the dug4 complementation group and hgt1 fall into the same complementation group (data not shown).
Identification of the DUG1 gene as YFR044c that encodes a protein belonging to the M20A metallohydrolase family:
To identify the gene corresponding to the dug1 mutant group, we transformed a dug1 mutant with a multicopy genomic library. Transformants were directly selected on SD–LEU + 0.2 mm GSH. Of an estimated 39,200 transformants, 61 grew on GSH. Complementation by MET15 would also lead to growth, as such transformants would be able to utilize inorganic sulfate. Therefore, all the transformants were plated on SD–LEU plates without an added organic sulfur source. Of 61 transformants, 51 grew on organic sulfur-free medium and were likely to be MET15 and were excluded from further analysis. Plasmids were extracted from 2 of the 10 yeast transformants that appeared to have plasmids other than MET15 and these were transformed into Escherichia coli.
Plasmids were isolated from these E. coli transformants and restriction fragment digestion was carried out to indicate unique plasmids. Reconfirmation of these plasmids was carried out by retransformation into yeast met15Δecm38Δdug1 and complementation of the growth defect on GSH was confirmed. The two purified plasmids that showed different restriction fragment patterns (pDUG1-C1 and pDUG1-C2) and were able to complement GSH growth defect were selected. One of the plasmids (pDUG1-C1) was subjected to sequence analysis by sequencing the two ends of the inserts. Sequencing and restriction mapping of the clone revealed an insert of ∼5.5 kb. The 5.5-kb insert contained two partial ORFs, YFR041c and YFR045w, and three complete ORFs, YFR042w, YFR043c, and YFR044c. All these ORFs are listed in the Saccharomyces Genome Database (SGD) as hypothetical uncharacterized ORFs of unknown function.
The protein sequences of the three ORFs YFR042w, YFR043c, and YFR044c, which were found in the clone that complemented the GSH growth defect in the dug1 mutant, were subjected to BLAST search to find homologs of known function across other organisms. The YFR042w gene encodes a protein of unknown function that is required for cell viability; YFRO43c encodes a protein with homology to proteins of unknown function in other organisms, while YFR044c encodes a protein that is homologous to a number of nonspecific peptidase homologs in both eukaryotes and prokaryotes. As Yfr044cp appeared to be the most likely candidate to complement the GSH utilization defect of the dug1 mutant, a met15Δyfr044cΔ strain was evaluated for growth on glutathione. The strain displayed an inability to grow on GSH, although it grew well on methionine- and cysteine-containing plates, and indicated that the dug1 locus carries mutations in YFR044c gene.
To further confirm that YFR044c alone could complement the defect of dug1, we cloned the YFR044c gene. The full gene was also cloned along with 600 bp of its native promoter to yield pRS313-YFR044c. pRS313-YFR044c restored the GSH utilization defect of the dug1 mutant while the cells transformed with vector alone showed no growth.
Sequence analysis of Yfr044cp predicted a protein of 481 amino acids. BLAST analysis of Yfr044cp proteins revealed a number of peptidase homologs in both eukaryotes and prokaryotes. The sequence similarity was highest to the M20A family of metallohydrolases, which belongs to the acylase-1 superfamily. This family includes glutamate carboxypeptidases, cytosolic nonspecific dipeptidases, carnosinases, aminopeptidases, and aminoacylases, each of which are characterized by their ability to hydrolyze different amide bonds (Figure 1B).
Identification of the DUG2 gene as YBR281c, which encodes a protein with an N-terminal WD40 repeat region and a C-terminal region showing homology to the M20A metallohydrolase family:
The DUG2 gene was cloned by complementation of the dug2 mutant as described for DUG1 above. Of the estimated 60,000 transformants, 55 passed the initial test of growth on GSH, of which 44 grew on organic sulfur-free medium and were picked for further analysis. Plasmids were extracted from 2 of the 11 remaining yeast transformants and transformed into E. coli, and unique plasmids were identified by restriction digestion. Reconfirmation was carried out by retransformation into yeast met15Δecm38Δdug2 and complementation of the growth defect on GSH was confirmed. The two purified plasmids that showed different restriction fragment patterns (pDUG2-C1 and pDUG2-C2) and were able to complement the GSH growth defect were selected. One of the plasmids (pDUG2-C1) was subjected to sequence analysis by sequencing the two ends of the inserts. Sequencing and restriction mapping of the clone revealed an insert of ∼6.235 kb. The insert contained two partial ORFs, YBR280c and YBR283c, and two complete ORFs, YBR281c and YBR282w. Analysis of ybr281cΔ deletions in a met15Δ background revealed a glutathione utilization defect, suggesting that YBR281c was the DUG2 gene. This was confirmed by cloning the YBR281c gene downstream of a translation elongation factor (TEF) 1α gene promoter and demonstrating that it complements the dug2 mutant defect of glutathione utilization.
Sequence analysis of Ybr281cp predicted a protein of 878 amino acids. BLAST analysis of the Ybr281cp protein reveals that the protein has two distinct domains. At the N-terminal domain (19–336 aa) is a putative WD40 repeat region, while the C-terminal region (423–878 aa) shows homology to the M20A metallopeptidase family (Figure 1C).
The WD40 sequence-containing repeat is a protein interaction motif that is normally present as a tandemly repeated unit of variable number. The sequence repeat unit is normally characterized by a Gly-His pair at the beginning of the repeat that ends with a Trp-Asp pair ∼40 amino acids downstream. The WD40 repeat region usually contains at least four repeat units. A minimum of four repeat units is required for folding into the higher-order β-propeller structure. The proteins containing the WD40 repeat regions participate in numerous functions that include cytoskeletal assembly, vesicular trafficking, signal transduction, control of transcription initiation complex, and mRNA splicing among others. The WD40 repeat region of these proteins is a modular interaction domain that is often a component of larger proteins and may function to form large multivalent complexes or act as a binding site for two or more proteins to enable transient interactions among these and other proteins to occur (Smith et al. 1999).
The Ybr281cp C-terminal region shows a significant similarity (33% identity and 51% similarity over a 368-amino-acid stretch) to Yfr044cp (Dug1p) and also belongs to the M20A metallohydrolases of the MEROPS (a database of peptidases) metallohydrolase clan of metallopeptidases (Rawlings and Barrett 1995). The Lactobacillus delbrueckii PepV dipeptidase, a member of the above-mentioned family whose crystal structure has been solved, and key amino acid residues involved in zinc binding, catalytic residue, and substrate binding have been identified (Jozic et al. 2002). The sequence alignment of the Ybr281cp (421–878 aa), Yfr044cp (1–481 aa), and L. delbrueckii PepV (1–470 aa) proteins reveals that the key zinc binding residues and the catalytic glutamic acid residue present in LdPepV are highly conserved across the three peptidases, although the residues important for substrate binding are not conserved (supplemental Figure 1 at http://www.genetics.org/supplemental/).
Identification of the DUG3 gene as YNL191w that encodes a protein belonging to the glutamine amidotransferase protein:
The DUG3 gene was cloned by complementation of the dug3 mutant as described for DUG1 above. Of the estimated 20,000 transformants, 62 passed the initial test of growth on GSH, of which 49 that grew on organic sulfur-free medium were excluded from further analysis. Plasmids were extracted from 2 of the 13 remaining yeast transformants and transformed into E. coli, and unique plasmids were identified by restriction digestion. Plasmids were isolated from these E. coli transformants and restriction fragment digestion was carried out to indicate unique plasmids. Reconfirmation of these plasmids was carried out by retransformation into yeast met15Δecm38Δdug3 and complementation of the growth defect on GSH was confirmed. The two purified plasmids that showed different restriction fragment patterns (pDUG3-C1 and pDUG3-C2) and were able to complement the GSH growth defect were selected. One of the plasmids (pDUG3-C1) was subjected to sequence analysis by sequencing the two ends of the inserts. Sequencing and restriction mapping of the clone revealed an insert of ∼6.287 kb. The insert contained two partial ORFs, YNL189w and YNL192w, and two complete ORFs, YNL190w and YNL191w.
Analysis of ynl191wΔ in a met15Δ background revealed a glutathione utilization defect, suggesting that YBR281c was the DUG3 gene. This was confirmed by cloning the YNL191w gene downstream of a TEF promoter and demonstrating that it complements the dug3 mutant defect of glutathione utilization. Sequence analysis of YNL191w reveals that ORF encodes a protein of ∼357 amino acids that contains a glutamine amido transferase (GATase) class II domain at its N-terminal region (Figure 1D). Members of the GATase class II family are able to catalyze the removal of ammonia from glutamine and the transfer to a substrate to form a new carbon–nitrogen bond. The family includes glucosamine-fructose-6-phosphate amido transferase, glutamine phosphorinbosyl pyrophosphate transferase, and asparagine synthase. This family of proteins (which have low sequence identity among themselves) is a member of the superfamily of N-terminal nucleophile (NTN) hydrolase families since the precursor proteins are usually synthesized with an N-terminal 11-residue propeptide that is cleaved off autocatalytically. The precursor protein is thus referred to as a peptidase (the mature protein, in contrast, lacks peptide-bond hydrolytic activity). The cys-12 residue, the nucleophile in the cleavage of the propeptide, becomes the N-terminal active site residue. In some members of this family, which appears to include Ynl191wp, the catalytic cysteine immediately follows the methionine start codon (Cys2) and is activated by cleavage of the N-terminal methionine by a methionine peptidase. As these precursors lack the autocatalytic peptidase activity, they are referred to as nonpeptidase homologs of the NTN hydrolase superfamily (Rawlings et al. 2006).
Sequence alignment study of Ynl191wp along with well-characterized Gln-AT-II enzymes was also performed. The Ynl191wp sequence was aligned with S. cerevisiae Gfa1p (glucoseamine-fructose 6-phosphate aminotransferase) (Milewski 2002), S. cerevisiae Ade4p (glutamine phosphoribosyl pyrophosphate amidotransferase), and E. coli PurF (amidophosphoribosyltransferase) (Muchmore et al. 1998). These proteins display low sequence similarity to each other. Thus the E. coli PurF, an amidophosphoribosyltransferase shows 27% identity (41% similarity) to Gfa1p over a 159-aa stretch and 25% identity (40% similarity) to Dug3p over a 108-aa stretch. More importantly, the amino acid residues, which are believed to play a crucial role in Gn_AT activity on the basis of the crystal structure, were also found to be conserved in Dug3p. These residues are the C2, R40, N121, G122, D151, R96, and T99 residues of Ynl191wp (supplemental Figure 2 at http://www.genetics.org/supplemental/). On the basis of the knowledge of the proteins belonging to this family, C2 is possibly the active site residue that may function as a nucleophile and attack the carboxyamide of glutamate and generate a γ-glutamyl thioester intermediate. N121 and G122 thereafter may play a crucial role in the formation of an oxyanion hole for the stability of the cysteinyl-glutamine tetrahedral intermediate. D151 and R96 side chains may provide the specificity for the α-amino and α-carboxyl groups of glutamine (Muchmore et al. 1998).
Analysis of extracts of dug1, dug2, and dug3 mutants for accumulation of GSH degradation intermediates:
Owing to the involvement of three new proteins in the alternative pathway of glutathione degradation, we attempted to determine the sequence of events by which these proteins may participate in the pathway by examining the different mutants for possible intermediates that may accumulate in these cells when grown in the presence of glutathione. This was carried out by growing the dug1, dug2, and dug3 mutant cells in methionine and then washing the cells and subsequently growing them in glutathione-containing medium for several hours. We observed maximal disappearance of glutathione from the medium after 16 hr of growth and this condition was chosen for evaluation of the extracts for novel thiol-containing intermediates. Cells were harvested, lysed, and deproteinized and equal amounts of extracts were treated with monobromobimane, which forms a fluorescent conjugate with glutathione and other thiol compounds containing a free cysteine, and evaluated using HPLC with a methanol/acetic acid solvent system as described in materials and methods. Although peaks from the control extracts corresponding to glutathione, γ-glu-cys, and cys-gly were detected, no accumulation of these compounds or any other novel peaks could be detected in the profiles of all three mutant extracts. The presence of a reducing agent, DTT, in the extracts did not affect the profiles significantly (data not shown). To eliminate the possibility that the accumulating thiols might be leaching out into the medium, we also examined the supernatant, but no new peaks could be detected in the supernatants.
Dug1p, Dug2p, and Dug3p are all required for the utilization of γ-glu-cys as a sole source of sulfur:
The γ-glu-cys dipeptide contains an unusual peptide bond similar to GSH. To examine how all the three Dug proteins participate in the degradation of GSH, the role of all these proteins in utilization of dipeptides like γ-glu-cys was also examined.
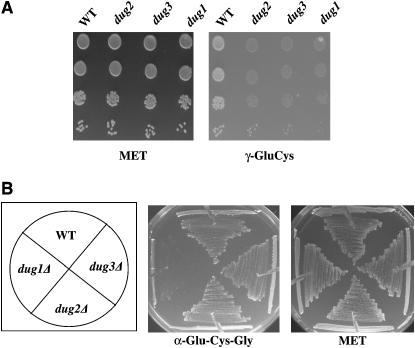
The strains met15Δecm38Δ, met15Δecm38Δdug1, met15Δecm38Δdug2, and met15Δecm38Δdug3 were grown and spotted onto plates containing methionine, GSH, and γ-glu-cys as the sole source of sulfur (at 0.2 mm concentration). It was observed that only met15Δecm38Δ grew on plates supplemented with γ-glu-cys. All the dug mutant strains showed growth defect and inability to utilize γ-glu-cys similar to their inability to utilize glutathione that also contains a γ-glu-cys bond (Figure 2A). To eliminate the possibility that γ-glu-cys was not being resynthesized to glutathione (by the enzyme glutathione synthase, encoded by GSH2 that had γ-glu-cys as a substrate), we created strains that were disrupted in both glutathione biosynthetic enzymes and the γ-glutamyl transpeptidase-encoding gene. This strain, ABC1664 (gsh1Δgsh2Δecm38Δmet15Δ), was evaluated for the ability to utilize γ-glu-cys as a sole source of sulfur. Trace amounts of glutathione (10 μm) were also added owing to the glutathione auxotrophy phenotype; however, these trace GSH levels were insufficient to meet the sulfur requirements (data not shown). These strains could use γ-glu-cys as a source of sulfur, suggesting that the utilization of γ-glu-cys occurred independently of both γ-GT and GSH2. The requirement of all three Dug proteins, namely Dug1p, Dug2p, and Dug3p, suggests that the γ-glu-cys is utilized via the alternative pathway.
Figure 2.—
Role of DUG1, DUG2, and DUG3 genes in the degradation and utilization of (A) γ-glu-cys and (B) α-glu-cys-gly. Growth of dug mutants and WT strain on SD media (A) supplemented with γ-glu-cys or methionine (control) at a concentration of 0.2 mm and (B) supplemented with α-glu-cys-gly or methionine (control) at a concentration of 0.2 mm.
Dug1p alone is required for the degradation and utilization of cys-gly dipeptide as a sole source of sulfur:
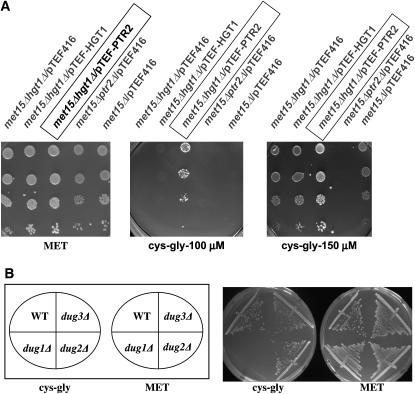
The possible role of Dug1p, Dug2p, and Dug3p in the utilization of cys-gly dipeptide (dipeptidyl moiety of GSH) as a sole source of sulfur was examined. However, when we examined the growth of met15Δ strains on cys-gly as a sole source of sulfur, we observed them to be very poorly utilized. We examined if the yeast dipeptide/tripeptide transporter Ptr2p or the glutathione transporter Hgt1p might be involved in the transport of this dieptide. As shown in Figure 3A, deletion of the PTR2 gene completely abolished transport of the cys-gly dipeptide, but deletion of the glutathione transporter HGT1 did not affect the transport of cys-gly. Even at higher concentrations along with longer incubations, no growth could be discerned in a ptr2Δ background although residual growth could otherwise be detected (Figure 3A). To increase the uptake of cys-gly and facilitate working with this dipeptide, the PTR2 gene was amplified by PCR and cloned downstream of the constitutive TEF promoter. Transformation of met15Δ strains with this pTEF-PTR2 plasmid permitted efficient utilization of cys-gly as a sulfur source (Figure 3A).
Figure 3.—
Utlilization of cys-gly as a sulfur source (A) Overexpression of PTR2 leads to enhanced transport of cys-gly. (B) Role of DUG1, DUG2, and DUG3 in utilization of cys-gly. (A) met15Δ hgt1Δ were transformed with control vectors pTEF416, pTEF-HGT1, and pTEF-PTR2, met15Δ and met15Δ ptr2Δ strains were transformed with control vector pTEF416, and serial dilution growth on methionine (0.2 mm), cys-gly dipeptide (0.10 mm), and cys-gly (0.15 mm) was measured. The plates were incubated for 2–3 days, except for the cys-gly (0.15 mm) plates, which were incubated for 7 days. (B) Growth of dug mutants and wild-type strains carrying a PTR2 (peptide transporter) overexpressing plasmid on cys-gly (0.2 mm)-containing medium.
Taking advantage of this, the strains met15Δ, met15Δdug1Δ, met15Δdug2Δ, and met15Δdug3Δ were transformed with the pTEF-PTR2 plasmid and growth was monitored on plates containing methionine or cys-gly as the sole organic source of sulfur. All the transformed strains grew equally well on methionine plates. None of the strains transformed with vector alone could grow on a plate containing cys-gly as the sole source of sulfur owing to poor utilization. However, in met15Δ strains bearing the pTEF-PTR2 plasmid, it was observed that the growth of met15Δdug2Δ and met15Δdug3Δ was comparable to that of met15Δ, indicating their ability to utilize cys-gly as a sulfur source. However, the met15Δdug1Δ strain bearing pTEF-PTR2 could not utilize cys-gly as the sole source of sulfur. This indicated that Dug1p, but not Dug2p or Dug3p, was required for the degradation and utilization of cys-gly as a source of sulfur (Figure 3B).
Dug1p alone is required for the degradation and utilization of α-glu-cys-gly tripeptide as the sole source of sulfur:
Studies with γ-glu-cys and cys-gly peptides revealed that Dug1p peptidase was required for the utilization of the cys-gly dipeptide. However, for the utilization of the γ-glu-cys dipeptide, Dug1p dipeptidase alone was not sufficient and the participation of the other proteins, Dug2p and Dug3p, were also required. This suggested that Dug1p might be able to act on dipeptides with a normal bond but not with an unusual peptide as presented in γ-glu-cys. To determine if Dug1p might also be able to utilize a tripeptide containing normal peptide bonds, the tripeptide α-glu-cys-gly was custom synthesized. The growth of the strains met15Δ, met15Δdug1Δ, met15Δdug2Δ, and met15Δdug3Δ were compared on plates containing either α-glu-cys-gly or γ-glu-cys-gly as a sulfur source. The α-glu-cys-gly peptide was efficiently utilized as a sulfur source by DUG+ met15Δ strains. Interestingly, while met15Δ dug2Δ and met15Δ dug3Δ strains grew well with this tripeptide as the sulfur source, the met15Δ dug1Δ strain failed to grow on plates containing α-glu-cys-gly (Figure 2B). This indicated that a functional Dug1p dipeptidase was required for the utilization of dipeptides or tripetides with a normal peptide bond; however, in dipeptides or tripeptides with unusual γ-glu linkages such as in γ-glu-cys-gly or in γ-glu-cys, all three proteins—Dug1p, Dug2p, and Dug3p—were required (Figures 1A and 2A).
Dug1p, Dug2p, and Dug3p interact to form a protein complex:
The inability to detect accumulation of any glutathione degradation intermediates in the dug mutants suggested that the three proteins might be participating in a multiprotein complex for the degradation of glutathione. Information at the SGD revealed that genomewide yeast two-hybrid studies had indicated interaction of Ybr281cp (Dug2p) with Ynl191wp (Dug3p) (Ito et al. 2001).
To determine if the proteins indeed interacted with each other, yeast two-hybrid analysis was carried out between the different proteins. We fused each of the Dug1p, Dug2p, and Dug3p proteins to the LexA DNA-binding protein in the yeast two-hybrid bait plasmid pEG202 and also to the GAL4 activation domain in the vector pJG4.5 as prey proteins using the yeast two-hybrid system DupLEX-A from Origene (Rockville, MD) based on the system developed by Gyuris et al. (1993). Two reporters, Leu2 (in the strain EGY48) and LacZ (in the plasmid pSH18-34) that were placed downstream of multiple copies of the LexA operator were used to evaluate the interaction (Gyuris et al. 1993). The bait fusions were also examined for autoactivation. Only the Bait-Dug2p showed mild autoactivation, and although we also used this fusion in some of the interactions, we ensured that the growth or X-Gal activity was above the activity observed by the autoactivation, and these putative interactions were also confirmed by the reverse fusions.
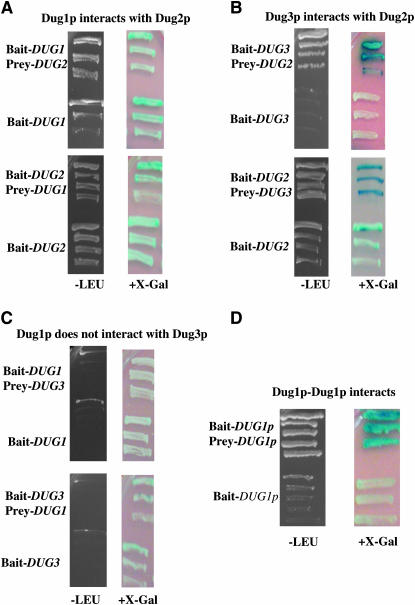
As can be seen from Figure 4A, Dug1p is capable of interacting with Dug2p in the interaction study in which we used pBait-Dug1 and pPrey-Dug2, as seen from the growth on −Leu plates as a reporter. With the LacZ reporter, the interaction was barely detectable over the background. However, in the reverse interaction where we used pBait-Dug2 and pPrey-Dug1, interactions over the background of autoactivation of pBait-Dug2 were not detectable owing to the autoactivation by pBait-Dug2 (Figure 4A). In the case of Dug2p and Dug3p, these proteins clearly interact with each other in both the forward and reverse interactions, once again using both the reporter readouts, growth on −Leu plates, and blue color formation on X-Gal plates (Figure 4B). Despite the autoactivation by pBait-Dug2p, in the presence of pPrey-Dug3 an increased activation above the background was observable. In the case of Dug1p interaction with Dug3p, however, Dug1p failed to interact directly with Dug3p as seen in both the forward and reverse interactions and in the reporter readouts, growth on −Leu plates, and blue color formation on X-Gal plates (Figure 4C). Taken together, the ability of Dug2p to interact with Dug1p and Dug3p also suggested the formation of a multimeric complex involving the three proteins.
Figure 4.—
Protein–protein interactions among Dug1p, Dug2p, and Dug3p. Yeast two-hybrid-based analyses where proteins are fused to the DNA-binding domain (Bait) or to the transcription activation domain (Prey). Positive interaction results either in growth on −LEU medium or in blue coloration on an X-gal plate.
The extensive yeast two-hybrid analysis interaction studies using both the forward and reverse Bait and Prey constructs and the dual reporter systems were strong indications of the interactions being true. However, as these can sometimes be artifactual, we also examined the possible interactions through co-immunoprecipitation (Co-IP) studies.
For the co-immunoprecipitation studies, we tagged the C terminus of all three Dug proteins differently. Tags were generated by a PCR-based method. The Dug1p was C-terminally tagged with either His or HA epitopes and cloned into p416TEF (URA3 marker) and pRS315 (LEU2 marker) or p414TEF (TRP1 marker). The Dug2p was C-terminally tagged with cMyc epitope and was cloned into p414TEF. The Dug3p was C-terminally tagged with HA epitope and was cloned into p416TEF. The tagged proteins were checked for their functionality using plate-based growth phenotype complementation of the respective mutants in a met15Δ background by examining growth on GSH. All the tagged proteins were found to be functional. The plasmids bearing the tagged genes were transformed into a protease-deficient strain (deficient in protease A, protease B, and carboxypeptidase Y), ABC 710, separately, in pairs, or all together, and cell extracts were prepared from these strains for further analysis.
To confirm the interaction between Dug2p and Dug3p, the Dug3pHA- and Dug2pcMyc-bearing plasmids were transformed together into the same yeast strain. Cell extracts were prepared and immunoprecipitation was done by using anti-cMyc antibody. Control samples contained only Dug2pcMyc-bearing plasmids and these cell extracts were also immunoprecipitated by using anti-cMyc antibody. The immunoprecipitation (IP) samples, as well as the total cell extract samples, were run on a denaturing gel, blotted, and probed with an anti-HA antibody. We could detect an ∼40-kDa band that corresponded to Dug3pHA in total cell extracts made from cells bearing both the plasmids and the corresponding IP sample. However, we could not detect Dug3pHA in the total cell extract of the control sample (containing only the Dug2p-cMyc plasmid) and its corresponding IP sample. This result indicates that Dug3pHA is co-immunoprecipitated along with Dug2pcMyc and suggests that Dug3pHA interacts with Dug2pcMyc (Figure 5A).
Figure 5.—
Co-immunoprecipitation assay to detect protein–protein interaction. (A) Co-immunoprecipitation of Dug2p and Dug3p. Tagged constructs of Dug2cMyc and Dug3HA were cotransformed or transformed along with vectors in ABC710. Cell extracts were prepared and immunoprecipation and immunoblotting were performed as described in materials and methods. Immunoprecipitation experiments were performed using anti-cMyc antibody (lanes 2, 4, and 5). The cell extracts of transformants as indicated were also loaded (lanes 1, 3, and 6). SDS–PAGE was followed with immunoblotting using anti-HA to detect Dug3HA. (B) Co-immunoprecipitation of Dug1p and Dug2p. Tagged constructs of Dug1His8 and Dug2cMyc were cotransformed or transformed along with vectors in ABC710. Cell extracts were prepared and immunoprecipation and immunoblotting were performed as described in materials and methods. Immunoprecipitation experiments were performed using anti-His antibody (lanes 2 and 4). The cell extracts of transformants as indicated were also loaded (lanes 1, 3, and 5). SDS–PAGE was followed with immunoblotting using anti-cMyc to detect Dug2cMyc. (C) Co-immunoprecipitation of the Dug1p homodimer. Tagged constructs of Dug1His8 and Dug1HA were cotransformed or transformed along with vectors in ABC710. Cell extracts were prepared and immunoprecipation and immunoblotting were performed as described in materials and methods. Immunoprecipitation experiments were performed using anti-His antibody (lanes 1 and 5). The cell extracts of transformants as indicated were also loaded (lanes 2, 3, and 6). SDS–PAGE was followed with immunoblotting using anti-HA to detect Dug1HA.
To examine Dug1p and Dug2p interaction, Dug1pHis8- and Dug2pcMyc-bearing plasmids were transformed together into the same yeast, and cell extracts were prepared. Immunoprecipitation was carried out using the anti-His antibody. Control cells for these experiments contained only Dug1pHis8 and these cell extracts were also immunoprecipitated by using anti-His antibody. Subsequently, the IP samples and the total cell extracts were run on a denaturing gel and probed using an anti-cMyc antibody. We could detect an ∼98-kDa band that corresponded to the Dug2pcMyc protein in total cell extracts made from cells containing both the plasmids and the corresponding IP samples of these extracts. However, we could not detect a corresponding band of 98-kDa Dug2pcMyc in the total cell extract of control samples bearing only the Dug1pHis8 plasmid and its corresponding IP sample. The result indicates that Dug2pcMyc co-immunoprecipitated along with Dug1pHis8 and suggests that Dug1pHis8 interacts with Dug2pcMyc (Figure 5B).
The Dug2p–Dug3p and Dug1p–Dug2p interactions seen in both the yeast two-hybrid analysis and the immunoprecipitation experiments and the presence of the WD40 domain in Dug2p, a domain known to play a role in many proteins in multiprotein interactions, suggested that perhaps these three proteins were forming a complex. To examine if they were indeed forming a protein complex involving all three proteins, double co-immunoprecipitation experiments were carried out. These experiments were carried out with yeast extracts made from transformants bearing all three plasmids corresponding to Dug1pHis8, Dug2pcMyc, and Dug3pHA. Immunoprecipitation was carried out on these cell extracts using anti-His antibody. The IP samples from these extracts were run on a denaturing gel electrophoresis, blotted, and probed separately with anti-HA (Figure 6A) and anti-cMyc antibody (Figure 6B). In these immunoblots, we could detect the ∼98-kDa Dug2pcMyc protein as well as the ∼40-kDa Dug3pHA protein both in the total cell extract where all three Dug proteins were coexpressed and in the corresponding IP sample (Figure 6, A and B). However, these bands could not be detected in control extracts, which were made from cells lacking either the Dug2p-cMyc plasmid (but containing the other two plasmids) or the Dug1p-HA plasmid (but containing the other two plasmids) (Figure 6, A and B).
Figure 6.—
Co-immunoprecipitation-based studies of Dug1p, Dug2p, and Dug3p reveal the formation of a ternary complex. Cell extracts from ABC710 transformants bearing all three plasmids (or with single or dual plasmids) containing different tagged DUG genes were first immunoprecipitated with anti-His and then detected with either (A) anti-cMyc or (B) anti-HA. Prior to addition of the first antibody, total cell extracts were also loaded along with other control extracts as indicated. Similar cell extracts were also first immunoprecipitated with anti-HA antibody and then detected with either (C) anti-His or (D) anti-cMyc antibody.
For further confirmation, the experiment was repeated using the same cell extracts, but by carrying out the first immunoprecipitation using anti-HA antibody. Subsequently, the immunoblots of all the samples—both the total cell extract and the IP samples—were probed using either anti-His (Figure 6C) or anti-cMyc antibodies (Figure 6D). We could detect Dug2pcMyc (∼98 kDa) as well as Dug1pHis8 (∼54 kDa) in the total cell extract of yeast samples where all three proteins and their corresponding IP samples were coexpressed (Figure 6, C and D). However, these bands could not be detected in control extracts, which were made from cells lacking either the Dug2p-cMyc plasmid (but containing the other two plasmids) or the Dug1p-His8 plasmid (but containing the other two plasmids) (Figure 6, C and D).
The result of the first Co-IP revealed that Dug3pHA and Dug2pcMyc co-immunoprecipitated along with Dug1pHis8. The result of the second Co-IP showed that Dug1pHis8 and Dug2pcMyc co-immunoprecipitated along with Dug3pHA. This suggests that Dug1pHis8, Dug2pcMyc, and Dug3pHA interact with each other to form a hetero-trimeric complex.
Protein interaction studies reveal that Dug1p can interact with itself:
We also examined possible Dug1p interaction with itself since some of the metallopeptidase M20A family members are known to dimerize. This was initially examined using the yeast two-hybrid analyses in which Dug1p fused to the LexA DNA-binding domain as bait protein and Dug1p fused to the GAL4 activation domain were examined for interaction. Using both the LEU2 reporter and the X-Gal reporter, it was clear that Dug1p could interact with itself (Figure 4D). These initial studies were further substantiated by immunoprecipitation studies. Dug1pHis8- and Dug1pHA-bearing plasmids were transformed into yeast cells, extracts were prepared, and immunoprecipitation was done by using anti-His antibody. Control yeast samples contained only Dug1pHis8-bearing plasmids, and the cell extract was immunoprecipitated by using anti-His antibody. Subsequently, immunoblots of all the samples, both total cell extract and IP samples, were detected by using anti-HA antibody. We could detect (∼54 kDa) Dug1HAp in the total cell extract of the test sample and its corresponding IP sample. However, we could not detect Dug1pHA in the total cell extract of the control sample and its corresponding IP sample. The result indicates that Dug1p interacts with itself (Figure 5D).
Dug1p, Dug2p, and Dug3p localize to the cytoplasm:
To obtain information on the possible location of Dug1p, Dug2p, and Dug3p proteins, we carried out studies using met15Δ yeast strains where the Dug1p, Dug2p, and Dug3p proteins were previously tagged with GFP at C-terminal ends, which were obtained from Invitrogen (San Diego) (Huh et al. 2003). The tagged proteins did not affect the function of Dug1p and Dug3p to any significant extent as seen by growth in glutathione as a sulfur source; however, a partial defect was seen in the Dug2p-GFP fusion strain. This strain was grown in methionine followed by reinoculation and incubation in GSH containing broth for several hours. Using confocal microscopy, all three proteins were observed to have uniform GFP fluorescence throughout the cytoplasm, which suggested that all three proteins were localized to the cytoplasm (supplemental Figure 3 at http://www.genetics.org/supplemental/).
DISCUSSION
In this article we have described a novel protein complex formed by the interaction of proteins encoded by three previously uncharacterized ORFs of S. cerevisiae (YFR044c, YBR281c, and YNL191w) that function specifically in glutathione degradation. The γ-glutamyl cycle, which describes the fates of glutathione (Orlowski and Meister 1970) and has remained essentially unchanged for >3 decades, thus has a new branch point in the degradation route, in addition to γ-glutamyl transpeptidase, which involves the proteins encoded by these ORFs in the form of a complex.
On the basis of the bioinformatic analysis, the physiological growth experiments of different strains bearing deletions in these genes, and the protein interaction studies, we propose a model for the functioning of the Dug1p, Dug2p, and Dug3p proteins in both peptide and glutathione degradation that is described below.
The Dug1p peptidase appears to be a nonspecific peptidase both from sequence alignments and from the experimental observations that only this protein (among the three) is required for the degradation of both the cysteinyl-glycine dipeptide and the α-glutamyl-cysteinyl-glycine tripeptide. As Dug2p and Dug3p do not participate in the degradation of these peptides, Dug1p is likely to have an activity independent of Dug2p and Dug3p. The members of the Dug1p peptidase family also have been found to have a predicted dimerization domain (Rawlings et al. 2006), and as interaction studies have shown that Dug1p can interact with itself, Dug1p is likely to form a homodimer for the degradation of di- and tripeptides. The specificity of this dipeptidase is not clear at this point, but, on the basis of its homology to other nonspecific dipeptidases, we predict that it would have a significant nonspecificity.
In the degradation of GSH or the dipeptide γ-glu-cys (and probably other di-/tripeptides containing a γ-glu-X bond), all three proteins—Dug1p, Dug2p, and Dug3p—are required. Under conditions where glutathione or compounds such as γ-glu-cys need to be degraded, it is likely that these three Dug proteins are brought together to form a complex, which recognizes GSH as a substrate and subsequently degrades it into its constituent amino acids. The ability of Dug2p (which contains both the WD40 domain and the peptidase domain) to interact with both Dug1p and Dug3p and the failure of Dug1p to interact with Dug3p suggests a likely model in which Dug3p interacts with Dug2p through the WD40 domain of Dug2p, while the M20 peptidase domain of the Dug2p that also contains a dimerization domain may allow the Dug1p-Dug2p proteins to interact. The presence of all the critical catalytic as well as the critical Zn2+-binding residues not only in Dug1p but also in Dug2 peptidase suggests that the Dug2p peptidase also plays a catalytic role in this degradation complex and does not function merely as a scaffolding protein.
What might be the function of Dug3p in this complex? Sequence analyses of Dug3p have shown the presence of a glutamine amidotransferase (Gln_AT) class II domain at the N terminus. The Gln_AT class II proteins like PurF and GlmS have been found to transfer the amine group of glutamine to an acceptor substrate. Dug3p could thus be involved in the amidation of γ-glutamyl residue of GSH. The likelihood of such a catalytic activity of Dug3p is supported by the observation that the key catalytic residues conserved in this class of proteins are also present in Dug3p. Dug3p may thus cleave and transfer the amide nitrogen of glutamine to the γ-glutamyl moiety of GSH, thereby modifying the γ-glutamyl residue, which then becomes recognizable by the Dug1p-Dug2p peptidase for subsequent peptidase action. Although, on the basis of present knowledge of the activity of various Gln_AT class II enzymes, multiple reaction schemes all involving amidation could be envisaged, the precise mechanism and modification by amidation would have to await experimental verification by in vitro assays, once they become possible.
GSH is present in almost all eukaryotes and even in some prokaryotes and is metabolized by the γ-glutamyl cycle. The possibility that this alternative pathway exists in organisms other than S. cerevisiae was investigated by searching databases for the presence of the homologs of Dug1p, Dug2p, and Dug3p in other organisms. Dug1p homologs were present in bacteria, fungi, and eukaryotes, including plants and mammals (the human homologs include cytosolic nonspecific dipeptidase and carnosinases). True orthologs of Dug2p or Dug3p, however, seemed to be restricted to the yeasts and fungi since similarities to proteins in other systems were restricted to the dipeptidase domain of Dug2p or to the glutamine amidotransferase domain of Dug3p (data not shown). These findings suggest that only some yeast and fungi have evolved a specialized mechanism of degradation of GSH independently of γ-GT, involving Dug1p, Dug2p, and Dug3p, and this mechanism is probably absent in bacteria, plants, and higher eukaryotes.
The acute glutathione toxicity seen in strains overexpressing the glutathione transporter Hgt1p (Srikanth et al. 2005) was not increased to any discernible extent as seen from the spot dilution assay when the alternative pathway was disrupted alone or along with the classical γ-GT pathway (data not shown). These results seem to suggest that both the alternative pathway and the γ-GT pathway might play a very minor role, if any, in dealing with excess glutathione levels in the cell.
In conclusion, the results described here on a novel protein complex mediating the degradation of glutathione have been arrived at through a combination of genetic, molecular, and physiological experiments. However, we have not yet been able to assess the activity of the protein complex in vitro. We are currently attempting to develop in vitro assays to demonstrate the degradation of glutathione in vitro that include the predicted amidation of the glutamyl residue by Dug3p. The development of in vitro biochemical assays and subsequent structural studies, as well as studies on the regulation of the proteins of this pathway, would ultimately lead us to greater insights into the function and mechanism of action of this novel degradation complex on this important metabolite.
Acknowledgments
We thank Raj Kumar for technical assistance, C. V. Srikanth and P. Vats for help with the PTR2 experiments, H. Kaur for help with the γ-glu-cys growth experiments and for help with some of the tag constructions, and R. Shaw for assistance with some of the yeast two-hybrid experiments. We thank Alok Mondal for help with the confocal microscope. C.K. and D.G. acknowledge financial support from Council of Scientific and Industrial Research, New Delhi, India. This work was partly funded by the Department of Science and Technology, Government of India.
References
- Aggarwal, M., and A. K. Mondal, 2006. Role of N-terminal hydrophobic region in modulating the subcellular localization and enzyme activity of the bisphosphate nucleotidase from Debaryomyces hansenii. Eukaryot. Cell 5: 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arrigo, A. P., 1999. Gene expression and the thiol redox state. Free Radic. Biol. Med. 27: 936–944. [DOI] [PubMed] [Google Scholar]
- Blaiseau, P. L., and D. Thomas, 1998. Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J. 17: 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, M. S., K. H. Johnston, G. J. Russell-Jones and E. C. Gotschlich, 1984. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal. Biochem. 136: 175–179. [DOI] [PubMed] [Google Scholar]
- Bourbouloux, A., P. Shahi, A. Chakladar, S. Delrot and A. K. Bachhawat, 2000. Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275: 13259–13265. [DOI] [PubMed] [Google Scholar]
- Brown, J. T., X. Bai and A. W. Johnson, 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, B., S. Ingavale and A. K. Bachhawat, 1997. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics 145: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey, R. C., and G. L. Newton, 1987. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 143: 85–96. [DOI] [PubMed] [Google Scholar]
- Fahey, R. C., and A. R. Sundquist, 1991. Evolution of glutathione metabolism. Adv. Enzymol. Relat. Areas Mol. Biol. 64: 1–53. [DOI] [PubMed] [Google Scholar]
- Fang, Y. Z., S. Yang and G. Wu, 2002. Free radicals, antioxidants, and nutrition. Nutrition 18: 872–879. [DOI] [PubMed] [Google Scholar]
- Ganguly, D., C. V. Srikanth, C. Kumar, P. Vats and A. K. Bachhawat, 2003. Why is glutathione (a tripeptide) synthesized by specific enzymes while TSH releasing hormone (TRH or thyroliberin), also a tripeptide, is produced as part of a prohormone protein? IUBMB Life 55: 553–554. [DOI] [PubMed] [Google Scholar]
- Grant, C. M., F. H. MacIver and I. W. Dawes, 1996. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 29: 511–515. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., E. Golemis, H. Chertkov and R. Brent, 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75: 791–803. [DOI] [PubMed] [Google Scholar]
- Hansen, J., and P. F. Johannesen, 2000. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 263: 535–542. [DOI] [PubMed] [Google Scholar]
- Heinemeyer, W., A. Simeon, H. H. Hirsch, H. H. Schiffer, U. Teichert et al., 1991. Lysosomal and non-lysosomal proteolysis in the eukaryotic cell: studies on yeast. Biochem. Soc. Trans. 19: 724–725. [DOI] [PubMed] [Google Scholar]
- Heinemeyer, W., A. Gruhler, V. Mohrle, Y. Mahe and D. H. Wolf, 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 26: 5115–5120. [PubMed] [Google Scholar]
- Heinemeyer, W., M. Fischer, T. Krimmer, U. Stachon and D. H. Wolf, 1997. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 272: 25200–25209. [DOI] [PubMed] [Google Scholar]
- Hilt, W., C. Enenkel, A. Gruhler, T. Singer and D. H. Wolf, 1993. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J. Biol. Chem. 268: 3479–3486. [PubMed] [Google Scholar]
- Holmgren, A., I. Ohlsson and M. L. Grankvist, 1978. Thiroedoxin from Escherichia coli. Radioimmunological and enzymatic determinations in wild type cells and mutants defective in phage T7 DNA replication. J Biol Chem. 253: 430–436. [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Hwang, C., A. J. Sinskey and H. F. Lodish, 1992. Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257: 1496–1502. [DOI] [PubMed] [Google Scholar]
- Ito, H., Y. Fukuda, K. Murata and A. Kimura, 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T., Chiba, R., Ozawa, M., Yoshida, M., Hattori, et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E. W., 2002. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351: 127–150. [DOI] [PubMed] [Google Scholar]
- Jozic, D., G. Bourenkow, H. Bartunik, H. Scholze, V. Dive et al., 2002. Crystal structure of the dinuclear zinc aminopeptidase PepV from Lactobacillus delbrueckii unravels its preference for dipeptides. Structure 10: 1097–1106. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis and A. Mitchell, 1994. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kumar, C., R. Sharma and A. K. Bachhawat, 2003. a Investigations into the polymorphisms at the ECM38 locus of two widely used Saccharomyces cerevisiae S288C strains, YPH499 and BY4742. Yeast 20: 857–863. [DOI] [PubMed] [Google Scholar]
- Kumar, C., R. Sharma and A. K. Bachhawat, 2003. b Utilization of glutathione as an exogenous sulfur source is independent of gamma-glutamyl transpeptidase in the yeast Saccharomyces cerevisiae: evidence for an alternative glutathione degradation pathway. FEMS Microbiol. Lett. 219: 187–194. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. W., 2002. Classical mutagenesis techniques. Methods Enzymol. 350: 189–199. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., and S. H. Bryant, 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi, K., and M. J. Penninckx, 1997. An important role for glutathione and gamma-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology 143: 1885–1889. [DOI] [PubMed] [Google Scholar]
- Meister, A., and M. E. Anderson, 1983. Glutathione. Annu. Rev. Biochem. 52: 711–760. [DOI] [PubMed] [Google Scholar]
- Milewski, S., 2002. Glucosamine-6-phosphate synthase: the multi-facets enzyme. Biochim. Biophys. Acta 1597: 173–192. [DOI] [PubMed] [Google Scholar]
- Muchmore, C. R., J. M. Krahn, J. H. Kim, H. Zalkin and J. L. Smith, 1998. Crystal structure of glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli. Protein Sci. 7: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski, M., and A. Meister, 1970. The gamma-glutamyl cycle: a possible transport system for amino acids. Proc. Natl. Acad. Sci. USA 67: 1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, W. C., S. N. Radyuk, L. Prabhudesai, D. Toroser, J. J. Benes et al., 2005. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J. Biol. Chem. 280: 37331–37338. [DOI] [PubMed] [Google Scholar]
- Ostergaard, H., C. Tachibana and J. R. Winther, 2004. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 166: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, M. J., and M. T. Elskens, 1993. Metabolism and functions of glutathione in micro-organisms. Adv. Microb. Physiol. 34: 239–301. [DOI] [PubMed] [Google Scholar]
- Perrone, G. G., C. M. Grant and I. W. Dawes, 2005. Genetic and environmental factors influencing glutathione homeostasis in Saccharomyces cerevisiae. Mol. Biol. Cell 16: 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N. D., and A. J., Barrett, 1995. Evolutionary families of metallopeptidases. Methods Enzymol. 248: 183–228. [DOI] [PubMed] [Google Scholar]
- Rawlings, N. D., F. R. Morton and A. J. Barrett, 2006. MEROPS: the peptidase database. Nucleic Acids Res. 34: D270–D272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Heiter, 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shi, Z. Z., J. Osei-Frimpong, G. Kala, S. V. Kala, R. J. Barrios et al., 2000. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc. Natl. Acad. Sci. USA 97: 5101–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T. F., C. Gaitatzes, K. Saxena and E. J. Neer, 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Srikanth, C. V., P. Vats, A. Bourbouloux, S. Delrot and A. K. Bachhawat, 2005. Multiple cis-regulatory elements and the yeast sulphur regulatory network are required for the regulation of the yeast glutathione transporter, Hgt1p. Curr. Genet. 47: 345–358. [DOI] [PubMed] [Google Scholar]
- Towbin, H., T. Staehelin and J. Gordon, 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wu, A. L., and W. S. Moye-Rowley, 1994. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14: 5832–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Y. Z. Fang, S. Yang, J. R. Lupton and N. D. Turner, 2004. Glutathione metabolism and its implications for health. J. Nutr. 134: 489–492. [DOI] [PubMed] [Google Scholar]