Abstract
ABC transporters constitute one of the largest gene families in all species. They are mostly involved in transport of substrates across membranes. We have previously demonstrated that the Caenorhabditis elegans ABC family shows poor one-to-one gene orthology with other distant model organisms. To address the evolution dynamics of this gene family among closely related species, we carried out a comparative analysis of the ABC family among the three nematode species C. elegans, C. briggsae, and C. remanei. In contrast to the previous observations, the majority of ABC genes in the three species were found in orthologous trios, including many tandemly duplicated ABC genes, indicating that the gene duplication took place before speciation. Species-specific expansions of ABC members are rare and mostly observed in subfamilies A and B. C. briggsae and C. remanei orthologous ABC genes tend to cluster on trees, with those of C. elegans as an outgroup, consistent with their proposed species phylogeny. Comparison of intron/exon structures of the highly conserved ABCE subfamily members also indicates a closer relationship between C. briggsae and C. remanei than between either of these species and C. elegans. A comparison between insect and mammalian species indicates lineage-specific duplications or deletions of ABC genes, while the family size remains relatively constant. Sites undergoing positive selection within subfamily D, which are implicated in very-long-chain fatty acid transport, were identified. The evolution of these sites might be driven by the changes in food source with time.
ATP-binding cassette (ABC) transporter proteins constitute one of the largest protein families in both prokaryotes and eukaryotes. In bacteria, they are involved primarily in the import of various sugars, vitamins, and amino acids into the cell. In eukaryotes, the majority of ABC transporter proteins are involved in exporting compounds across cytoplasmic membranes or into intracellular compartments such as the endoplasmic reticulum or mitochondria. No eukaryotic ABC transporter has been found to be involved in import of compounds from outside the cell (Saurin et al. 1999). A typical ABC transporter consists of at least one evolutionarily conserved ABC domain (also known as the nucleotide-binding domain, or the NBD), comprising ∼200 amino acid residues and a transmembrane domain (TMD) containing several predicted transmembrane α-helices. An ABC domain usually contains a Walker A and Walker B motif, which are also found in other nucleotide-binding proteins, and an ABC signature (C) motif, located just upstream from the Walker B site. The C motif usually contains the consensus sequence LSGGQK, which is diagnostic of ABC transporters and distinguishes them from other ATPases. The eukaryotic ABC genes are organized either as full transporters containing two TMDs and two ABCs or as half transporters containing one of each (Hyde et al. 1990). The half transporters are thought to form either homodimers or heterodimers to form a functional transporter.
We previously reported 60 ABC genes in Caenorhabditis elegans and classified them into eight subfamilies, A–H, on the basis of amino acid sequence and domain organization. One ABC gene, Y74C10AR.3, was missed in our previous analysis but is included here for comparative analysis. A phylogenetic analysis of the ABC genes of C. elegans and three non-nematode eukaryotic species indicated that the level of orthology is substantially lower than was expected (Sheps et al. 2004). Release of the genome sequences of both C. briggsae (Stein et al. 2003) and C. remanei (http://genome.wustl.edu) enables us to examine the evolution dynamics of the ABC family among more closely related species.
Gene duplication is an important source for the evolution of gene diversity. Several hypotheses have been proposed regarding the fate of the duplicated copies: (1) one duplicated paralog is under neutral selection and becomes a pseudogene with time (nonfunctionalization by degeneration); (2) one paralog adopts a new function following advantageous mutation (neofunctionalization by positive/adaptive selection); or (3) the original functions are partitioned between the two duplicated copies (subfunctionalization) (Lynch and Conery 2000). It is striking that about half of the membrane protein-encoding genes in many sequenced genomes were found to be within tandem clusters (Kihara and Kanehisa 2000). C. elegans ABC genes are also frequently observed in tandem clusters. Expression analysis of these locally duplicated ABC genes suggested that many of them may be subject to subfunctionalization (Zhao et al. 2004). While orthology is rare between ABC genes of C. elegans and distant eukaryotic organisms, the total number of ABC genes in each is about the same, implying that ABC genes underwent species-specific duplications and losses (Sheps et al. 2004). Given that the subfunctionalization hypothesis provides a plausible explanation for retaining tandemly duplicated ABC genes, what selection pressures drive the evolution of closely related paralogous ABC genes within a given subfamily as a whole? To address this question, we examined whether positive selection (adaptive evolution) plays a role in shaping ABC subfamily dynamics. Selective pressure at the protein level is usually measured by the nonsynonymous (dN)/synonymous (dS) rate ratio (ω). dN/dS is expected to be 1.0 for genes under neutral selection, <1.0 for genes under purifying selection, and >1.0 for genes under adaptive selection. Recently developed codon-based models take into account variations of the ratio among sites (Nielsen and Yang 1998; Yang et al. 2000). Detecting adaptive selection has played a critical role in understanding the mechanisms of molecular evolution of different gene families (Thomas et al. 2005). ABC proteins are able to transport an unusually broad range of substrates. Evaluation of adaptively selected sites among closely related members might provide insights into the mechanisms of substrate recognition by ABC proteins.
MATERIALS AND METHODS
Prediction of ABC genes in C. briggsae and C. remanei:
All 61 protein sequences of ABC transporters in C. elegans were retrieved from WormBase (WS160) (Table 1). To identify putative C. briggsae and C. remanei ABC genes, a single protein sequence from each C. elegans subfamily was used as a query against C. briggsae genomic sequence (cb25.agp8) or C. remanei draft sequence (C. remanei Pcap Assembly) by standalone TBLASTN (available from NCBI) using default parameters. Contig hits with an E-value <10−10 were pooled and redundant hits were removed. These contig sequences were subjected to ab initio gene prediction by FGENESH (Salamov and Solovyev 2000) and/or GenScan (Burge and Karlin 1997). The resulting protein sequences were used as query to scan Pfam (Bateman et al. 2002) to identify candidate ABC genes. Then these candidates were searched against WormPep (WS160) by BLASTP. If more than five of the top hits are ABC genes, the query is retained as an ABC gene. The putative ABC genes were further classified into eight different subfamilies as described previously (Sheps et al. 2004). The resulting C. briggsae ABC gene set has been integrated with those annotated in WS160. To refine ABC gene annotation in C. remanei, we have also used homology-based gene annotation programs, including GENEWISE (Birney et al. 2004) and EXONERATE (Slater and Birney 2005), to take the advantage of the high-quality annotation of its sister species, C. elegans. The C. remanei genome sequences have been assembled and made publicly available by the Washington University Genome Sequence Center (http://genome.wustl.edu/). Protein sequences coded by C. elegans ABC genes were taken from WormBase (WS160) (Chen et al. 2005). The annotation procedure consists of two major steps. First, C. remanei genomic sequences that encode candidate ABC genes by using C. elegans ABC genes as query to search the C. remanei genomic sequences database using WU-BLAST (tblastn) (Lopez et al. 2003). Regions that best match the query C. elegans ABC genes were parsed and recorded. Second, each C. elegans ABC protein and its corresponding C. remanei genomic region were fed to EXONERATE with the setting “-model protein2genome -n 1–refine full” and to GENEWISE with the setting “-gap 12 -e 8 -alg 623L.” Protein sequences and gene models are parsed and recorded for further analyses and display. The ABC gene set from FGENESH and GENEWISE predictions was finally subjected to substantial manual editing to correct improper predictions such as intron/exon boundaries, open reading frame (ORF) fusions, splitting, and missing exons. The genomic positions of putative ABC genes in C. briggsae and C. remanei are listed in supplemental Table 3 at http://www.genetics.org/supplemental/.
TABLE 1.
ABC genes in C. elegans, C. briggsae, and C. remanei
|
C. elegans
|
C. briggsae
|
C. remanei
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subfamily | Gene name | ORF name | Protein size | First exon EST | ORF namea | Protein size | Syntenyb | Inversion | ORF namea | Protein size | Syntenyb | Inversion |
| A (7)c | Abt-2 | F12B6.1 | 1531 | No | br|F12B6.1 | 1534 | Yes | No | rm|F12B6.1 | 1543 | Yes | Yes |
| Ced-7 | C48B4.4 | 1689 | Yes | br|C48B4.4 | 1694 | Yes | No | rm|C48B4.4 | 1688 | Yes | No | |
| Abt-3 | F55G11.9d | 1431 | No | — | — | — | — | rm|ABCA.1e | 1505 | No | NAf | |
| NAf | F56F4.6d | 260 | No | br|ABCA.1 | 1546 | Yes | No | rm|ABCA.2 | 1539 | Yes | No | |
| Abt-5 | Y53C10A.9d | 1564 | No | br|ABCA.2 | 1521 | No | NA | rm|ABCA.3 | 1981 | No | NA | |
| Abt-4 | Y39D8C.1 | 1802 | No | br|Y39D8C.1 | 1805 | No | NA | rm|Y39D8C.1 | 1801 | No | NA | |
| Abt-1 | C24F3.5 | 1429 | No | br|C24F3.5 | 1429 | Yes | No | rm|C24F3.5 | 1403 | Yes | No | |
| B (25) | Pgp-10 | C54D1.1 | 1283 | No | br|C54D1.1 | 1275 | Yes | No | rm|C54D1.1 | 1274 | Yes | No |
| Pgp-11 | DH11.3 | 1239 | No | br|DH11.3 | 1209 | Yes | No | rm|DH11.3 | 1178 | Yes | No | |
| Pgp-1 | K08E7.9 | 1321 | No | br|K08E7.9 | 1319 | Yes | Yes | rm|K08E7.9 | 1320 | Yes | Yes | |
| Pgp-9 | C47A10.1 | 1294 | No | br|C47A10.1 | 1294 | Yes | Yes | rm|C47A10.1 | 851g | Yes | Yes | |
| Pgp-2 | C34G6.4 | 1265 | No | br|C34G6.4 | 1265 | Yes | No | rm|C34G6.4 | 1265 | Yes | No | |
| Pgp-3 | ZK455.7d | 1268 | No | br|ZK455.7 | 1268 | Yes | No | rm|ZK455.7 | 1268 | Yes | No | |
| Pgp-4 | F42E11.1d | 1266 | Yes | br|F42E11.1 | 1265 | Yes | No | rm|F42E11.1 | 1226 | Yes | No | |
| Pgp-12 | F22E10.1 | 1318 | No | br|F22E10.1 | 1315 | Yes | No | rm|F22E10.1 | 1318 | Yes | No | |
| Pgp-13 | F22E10.2d | 1324 | No | br|F22E10.2 | 1323 | Yes | No | rm|F22E10.2 | 1323 | Yes | No | |
| Pgp-14 | F22E10.3d | 1327 | Yes | br|F22E10.3 | 1327 | Yes | Noh | rm|F22E10.3 | 1327 | Yes | No | |
| Pgp-15 | F22E10.4d | 1328 | No | br|F22E10.4 | 1331 | Yes | No | rm|F22E10.4 | 1330 | Yes | No | |
| Pgp-8 | T21E8.3d | 1238 | Yes | br|ABCB.3 | 1240 | Yes | Noh | rm|ABCB.3 | 1263 | Yes | No | |
| Pgp-7 | T21E8.2d | 1269 | No | br|ABCB.2 | 1254 | Yes | No | rm|ABCB.2 | 1263 | Yes | No | |
| Pgp-6 | T21E8.1d | 1264 | No | br|ABCB.1 | 1256 | Yes | No | rm|ABCB.1 | 1257 | Yes | No | |
| Pgp-5 | C05A9.1d | 1283 | No | — | — | — | — | — | — | — | — | |
| NA | Y74C10AR.3 | 704 | No | br|Y74C10AR.3 | 704 | Yes | No | rm|Y74C10AR.3 | 612g | No | No | |
| Haf-2 | F43E2.4 | 761 | Yes | br|F43E2.4 | 763 | Yes | No | rm|F43E2.4 | 761 | Yes | No | |
| Haf-4 | W04C9.1 | 787 | Yes | br|W04C9.1 | 787 | Yes | No | rm|W04C9.1 | 786 | Yes | No | |
| Haf-9 | ZK484.2 | 815 | Yes | br|ZK484.2 | 816 | Yes | No | rm|ZK484.2 | 815 | No | NA | |
| Haf-7 | Y50E8A.16 | 807 | Yes | br|Y50E8A.16 | 795 | Yes | No | rm|Y50E8A.16 | 801 | No | NA | |
| Haf-8 | Y57G11C.1 | 633 | No | — | — | — | — | — | — | — | — | |
| Haf-6 | Y48G8AL.11 | 565 | Yes | br|Y48GAL.11 | 563 | Yes | D/I | rm|Y48GAL.11 | 564 | Yes | No | |
| Haf-1 | C30H6.6 | 586 | No | br|C30H6.6 | 584 | Yes | Yes | rm|C30H6.6 | 584 | Yes | No | |
| Haf-3 | F57A10.3 | 733 | No | br|F57A10.3 | 706 | Yes | No | rm|F57A10.3 | 702 | Yes | No | |
| Haf-5 | W09D6.6 | 801 | Yes | br|W09D6.6 | 803 | Yes | D/Ii | rm|W09D6.6 | 798 | Yes | No | |
| C (9) | Mrp-5 | F14F4.3 | 1400 | Yes | br|F14F4.3 | 1397 | Yes | No | rm|F14F4.3 | 1395 | Yes | No |
| Mrp-6 | F20B6.3 | 1396 | No | br|F20B6.3 | 1415 | Yes | No | rm|F20B6.3 | 1412 | Yes | No | |
| Mrp-2 | F57C12.4 | 1525 | Yes | br|F57C12.4 | 1530 | Yes | No | rm|F57C12.4 | 1525 | Yes | No | |
| Mrp-1 | F57C12.5 | 1528 | Yes | br|F57C12.5 | 1524 | Yes | No | rm|F57C12.x | 1521 | Yes | No | |
| Mrp-3 | E03G2.2 | 1398 | No | br|E03G2.2 | 1399 | Yes | D/I | rm|E03G2.2 | 1379 | Yes | No | |
| Mrp-4 | F21G4.2 | 1573 | No | br|F21G4.2 | 1567 | Yes | No | rm|F21G4.2 | 1573 | Yes | No | |
| Mrp-7 | Y43F8C.12 | 1153 | No | br|Y43F8C.12j | 1003 | No | NA | rm|Y43F8C.12 | 1111 | Yes | No | |
| Mrp-8 | Y75B8A.26 | 1114 | Yes | br|Y75B8A.26 | 1125 | No | NA | rm|Y75B8A.26 | 1133 | Yes | No | |
| NA | C18C4.2 | 1247 | Yes | br|C18C4.2 | 1245 | Yes | D/I | rm|C18C4.2 | 1238 | Yes | No | |
| D (5) | pmp-5 | T10H9.5 | 598 | Yes | br|T10H9.5 | 598 | Yes | No | rm|T10H9.5 | 597 | Yes | No |
| pmp-3 | C54G10.3 | 660 | Yes | br|C54G10.3 | 660 | Yes | No | rm|C54G10.3 | 660 | Yes | No | |
| pmp-4 | T02D1.5 | 734 | Yes | br|T02D1.5 | 733 | Yes | No | rm|T02D1.5 | 733 | Yes | No | |
| pmp-1 | C44B7.8 | 665 | Yes | br|C44B7.8 | 663 | Yes | No | rm|C44B7.8 | 663 | Yes | No | |
| pmp-2 | C44B7.9 | 661 | Yes | br|C44B7.9 | 662 | Yes | No | rm|C44B7.9 | 662 | Yes | No | |
| E (1) | NA | Y39E4B.1 | 610 | Yes | br|Y39E4B.1 | 610 | Yes | No | rm|Y39E4B.1 | 610 | Yes | No |
| F (3) | NA | T27E9.7 | 622 | Yes | br|T27E9.7 | 620 | Yes | Yes | rm|T27E9.7 | 622 | Yes | No |
| NA | F42A10.1 | 712 | Yes | br|F42A10.1 | 712 | Yes | No | rm|F42A10.1 | 711 | Yes | No | |
| NA | F18E2.2 | 622 | Yes | br|F18E2.2 | 622 | Yes | No | rm|F18E2.2 | 622 | Yes | No | |
| G (9) | NA | Y42G9A.6 | 620 | No | br|Y42G9A.6 | 595 | Yes | No | rm|Y42G9A.6 | 642 | Yes | No |
| NA | C05D10.3 | 598 | Yes | br|C05D10.3 | 649 | Yes | No | rm|C05D10.3 | 597 | Yes | No | |
| NA | F02E11.1 | 658 | No | br|F02E11.1 | 569 | Yes | NA | rm|F02E11.1 | 567 | Yes | No | |
| NA | F19B6.4 | 695 | Yes | br|F19B6.4 | 654 | Yes | NA | rm|F19B6.4 | 644 | No | NA | |
| NA | Y49E10.9 | 454 | No | br|Y49E10.9 | 462 | No | NA | rm|Y49E10.9 | 452 | No | NA | |
| NA | Y47D3A.11 | 547 | No | br|Y47D3A.11 | 547 | Yes | No | rm|Y47D3A.11 | 547 | Yes | D/I | |
| NA | C16C10.12 | 610 | No | br|C16C10.12 | 599 | Yes | D/I | rm|C16C10.12 | 597 | Yes | D/I | |
| NA | C10C6.5 | 610 | Yes | br|C10C6.5 | 610 | Yes | No | rm|C10C6.5 | 610 | Yes | D/I | |
| NA | T26A5.1 | 608 | No | br|T26A5.1 | 613 | Yes | Yes | rm|T26A5.1 | 575 | Yes | Yes | |
| H (2) | NA | C56E6.1 | 1677 | No | br|C56E6.1 | 1666 | Yes | No | rm|C56E6.1 | 1665 | Yes | No |
| NA | C56E6.5 | 595 | Yes | br|C56E6.5 | 593 | Yes | No | rm|C56E6.5 | 595 | Yes | No | |
C. briggsae and C. remanei ABC genes are assigned the same ORF name as its C. elegans orthologs prefixed by “br|” and “rm|,” respectively.
See materials and methods.
Numbers in parentheses are the number of members in the subfamily for C. elegans.
C. briggsae orthologs not assigned in Wormbase (WS160).
Genes that are underlined are not orthologs for any single gene and their position in the table is arbitrary.
NA, not applicable.
Partial peptide sequences are due to incomplete sequences on the contig.
A transposase-like ORF was found between br|F22E10.2 and br|F22E10.3; a reverse transcriptase-like ORF was found between br|ABCB.1 and br|ABCB.2.
Deletion or insertion.
This gene is assembled from two contigs: cb25.NA_172 and cb25.NA_007. The first few exons could be missing because the gene starts at the very beginning of cb25.NA_172.
Ortholog assignment:
To assign C. briggsae or C. remanei orthologs for C. elegans ABC genes, a multiple alignment was generated for each subfamily using sequences from the three nematode species, humans, mice, and two insect species, Drosophila melanogaster and D. pseudoobscura. Alignment was done with CLUSTALW with gap distance=0 and matrix=BLOSUM but otherwise default settings (Higgins and Sharp 1988; Thompson et al. 1997). Regions with many gaps were removed from the multiple alignments using BONSAI (J. Thomas, personal communication). The resulting alignments were used to estimate a maximum-likelihood (ML) phylogenetic tree with 500 bootstraps using the PHYML program with the following parameters: JTT substitution matrix, six rate categories, gamma parameter 1.0, and no invariant sites (Guindon and Gascuel 2003). Genes were assigned as one-to-one orthologs when they formed a pure cluster with one gene from each species on the tree with strong bootstrap support. In these cases, the C. briggsae and C. remanei genes were named after C. elegans ABC genes prefixed by “br|” and “rm|,” respectively.
Synteny information was recorded by performing BLASTP searches against WormPep using predicted proteins from C. briggsae or C. remanei ABC candidates and flanking genes as query. Synteny information was used as a reference for genomic dynamics but not as a criterion for assignment of ortholog. If two of the three consecutive genes flanking either side of the gene of interest were found to be in the same order in both animals, then synteny was assumed. If the gene of interest was found in a different orientation in relation to its adjacent one compared with those in C. elegans, an inversion was assumed. If three consecutive genes flanking either side of the gene of interest in C. elegans were found missing in the syntenic region of C. briggsae and C. remanei, a deletion was assumed. All other cases such as ambiguity or lack of flanking sequences were recorded as not applicable (NA). C. remanei gene models of the candidate ABC genes were processed in GFF format (http://www.sanger.ac.uk/Software/formats/GFF/) and were then loaded into a Bio∷DB∷GFF database for visualization with a searchable generic genome browser (Stein et al. 2003). C. remanei ABC genes were searched and visualized for their genomic neighborhoods. The resulting syntenic data were incorporated with those from BLASTP searching of the flanking genes. ABC protein sequences for insect and mammalian species were retrieved from an ENSEMBL database based on a published ABC gene list (Dean et al. 2001; Sheps et al. 2004), except for D. pseudoobscura for which the ABC sequences were recovered from a BLASTP with default settings.
Intron alignment:
C. elegans Y39E4B.1 (ABCE) intron positions were retrieved from WormBase (WS 160). Intron positions of br|Y39E4B.1 and rm|Y39E4B.1 were derived from ab initio prediction of c002401148.Contig1 and Supercontig27.35, respectively, by FGENESH. To test the reliability of the prediction, a C. elegans genomic region encoding Y39E4B.1 was used as input for FGENESH. The predicted gene model was identical to the WormBase (WS160) annotated one confirmed by expressed sequence tags (ESTs), indicating the high accuracy of ABCE subfamily prediction by FGENESH (data not shown).
Identification of positive selection sites:
We analyzed seven data sets consisting of ABCA, full- and half-sized ABCB, ABCC, D, F, and G subfamilies. F56F4.5 was excluded from subfamily A because of its unusually small size. Subfamilies E and H were excluded because they contain too few genes to analyze (one and two each, respectively). For each data set, a protein alignment was produced by CLUSTALW as described above. A ML tree was estimated by PHYML. A cDNA alignment that corresponded to its amino acid alignment was prepared, and the trees and alignments were used as input to CODEML. Sites with gaps were excluded from the analysis. Three different initial ω-values (0.3, 1.0, and 3.4) were used to run CODEML for each data set using model 7 and 8. For cases in which the additional dN/dS ratio assigned by model 8 was >1, significance was tested by a χ2 test (with 2 d.f.) on twice the negative of the log-likelihood difference between models 7 and 8 (Thomas et al. 2005). Specific sites under positive selection were those with probability >0.85 as determined by Bayes Empirical Bayes (BEB) analysis (Yang et al. 2000). The predicted sites were mapped to the secondary structure of a representative ABC protein, with ABC and TMD predicted by Pfam (Bateman et al. 2002) and TMHMM (Krogh et al. 2001), respectively. The sites were also indicated by a star on the alignment produced in GeneDOC (Nicholas and Nicholas 1997).
Confirmation of ortholog assignment by promoter-driven green fluorescence protein expression:
We further tested whether ortholog assignments between C. elegans and C. briggsae were supported by functional analysis. A subset of ABC genes, especially those from tandem-duplicated ones, were chosen to verify whether a similar expression patterns can be observed. A fusion PCR technique was used to generate the promoter∷green fluorescence protein (GFP) construct as described (Hobert 2002). The fusion PCR product was co-injected with wild-type dpy-5 rescue DNA (pCeh361) (Thacker et al. 2006) into dpy-5 mutant CB907 with the concentration of 10 and 100 ng/μl, respectively. Visualization and imaging of GFP expression were done as described (Zhao et al. 2004).
Mutant manipulations:
Most of the single-gene mutant strains were generated by C. elegans Knockout Consortium. Outcrossing was performed at least twice with N2 Bristol male worms. None of them shows obvious phenotypes under normal growth conditions except for the T21E8.2 deletion mutant, which is lethal (supplemental Table 2 at http://www.genetics.org/supplemental/). The lethality is caused by mutations closely linked to T21E8.2 on the basis of the following observations: first, a T21E8.1 and -2 double mutant appears to be wild type; second, cosmid T21E8, which covers the full-length genomic DNA of T21E8.2, did not rescue the lethality (data not shown).
RESULTS
ABC genes are well conserved among C. elegans, C. briggsae, and C. remanei:
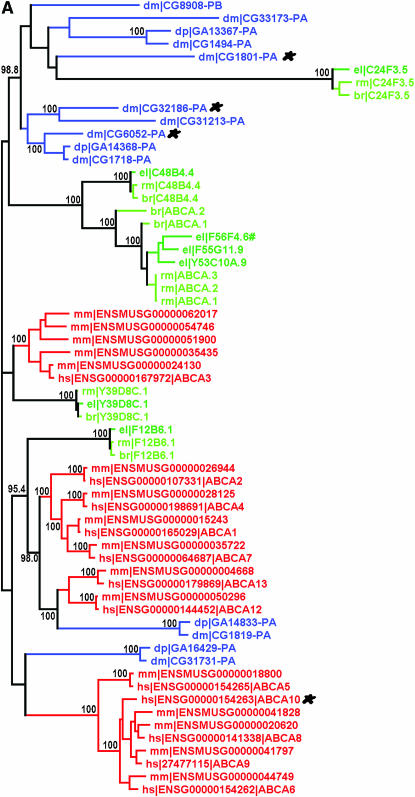
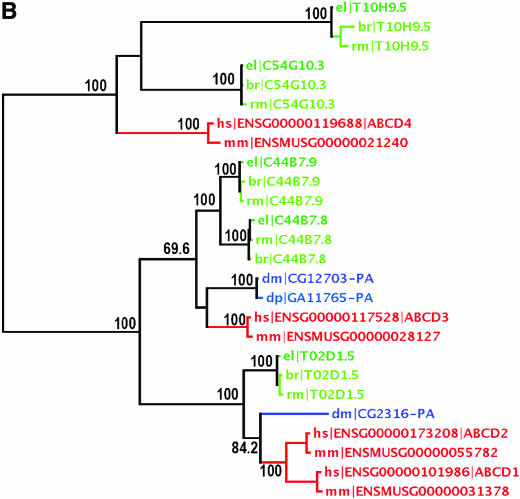
We reported 60 ABC genes in the C. elegans genome and grouped them into eight subfamilies according to their sequence similarity and domain organization (Sheps et al. 2004). Here we identified putative ABC genes in C. briggsae and C. remanei using a combination of ab initio gene prediction and database searching (see materials and methods). One of the C. elegans ABC genes, Y47C10AR.3, was missed in our previous report but included in this analysis. As a result, we were able to identify 58 and 59 ABC genes in the genomes of C. briggsae and C. remanei, respectively, which were grouped into subfamilies as described above. An ML phylogenetic tree was estimated for each subfamily using protein sequences from all three species (Figure 1, supplemental Figure 1 at http://www.genetics.org/supplemental/). To facilitate the comparison of the nematode ABC genes with those of other organisms, we also included ABC sequences from humans, mice, and two fly species, D. melanogaster and D. pseudoobscura. Given the substantial differences in domain content, subfamily B was subdivided into two categories of genes: full-sized (∼1400 amino acids on average) and half-sized (∼700 amino acids on average). A separate ML tree was estimated for each of these two categories (supplemental Figure 1, a and b, at http://www.genetics.org/supplemental/). Orthology describes genes separated from one another by speciation while paralogy describes those separated by gene duplication events (Fitch 1970). C. briggsae and C. remanei ABC genes were assigned as one-to-one C. elegans orthologs when they were present on a tree branch consisting of a single ABC gene in each species. They were named after the corresponding C. elegans gene prefixed by “br|” and “rm|,” respectively. C. elegans genes are prefixed by “el|” only in the phylogenetic trees for ease of tree interpretation. In situations where a single ABC gene or a set of duplicated ABC genes in one species are on a branch with a set of duplicated ones in another species, these genes form co-orthologous groups with one another (Sonnhammer and Koonin 2002). In these cases, the ABC genes in C. briggsae or C. remanei were named consecutively by subfamily prefixed by “br|” and “rm|,” respectively (Table 1; Figure 1; supplemental Figure 1 at http://www.genetics.org/supplemental/). In contrast to low orthology between ABC genes of C. elegans and other organisms, frequent one-to-one orthology was observed between ABC genes from the three nematode species. Similar levels of orthology were observed between humans and mice and between the two Drosophila species (Figure 1; supplemental Figure 1 at http://www.genetics.org/supplemental/). Of the 61 C. elegans ABC genes, 53 were found to have one-to-one orthologs in C. briggsae and C. remanei (Table 1). Nearly all of these orthologs are also found in syntenic regions in each genome. Species-specific expansion or loss of ABC genes was seen primarily in the ABCA and full-sized ABCB subfamilies (Figure 1A; supplemental Figure 1a at http://www.genetics.org/supplemental/). There are, in total, 16 tandemly duplicated ABC genes in the C. elegans genome, 10 of which are members of subfamily B (supplemental Figure 2 at http://www.genetics.org/supplemental/). C. briggsae or C. remanei orthologs were assigned for 6 of these 10 genes (Table 1), while only 1, F22E10.1, was assigned a C. briggsae ortholog previously (Stein et al. 2003). High levels of orthology were observed among the three nematode species for all the remaining subfamilies, including subfamily B (half-sized), C, D, E, F, G, and H (Figure 1B; supplemental Figure 1, b–g, at http://www.genetics.org/supplemental/). To test the functional significance of the ortholog assignments, we chose a few orthologous pairs between C. elegans and C. briggsae and compared their anatomical expression patterns. The putative promoter sequences from these pairs drove GFP expression in similar patterns in all instances (supplemental Table 1 at http://www.genetics.org/supplemental/). The conservation of promoter elements was further confirmed by similar expression patterns resulting from reciprocal introduction of promoter∷GFP constructs into each species, indicating that orthologous ABC genes between nematode species very likely share similar functions.
Figure 1.—
Maximum-likelihood protein trees of ABC genes from subfamily A (A) and subfamily D (B). The two subfamilies were chosen to show nematode ABC genes with or without species-specific clustering in the phylogenetic trees. Those genes in a clade consisting of one ABC gene from each nematode species are defined orthologs. Members from nematode, insect, and mammalian species are green, blue, and red, respectively. Numbers on the branch nodes indicate the percentage of bootstrap support from 500 replicates. Genes marked by stars probably have an unpredicted ortholog in the related species on the basis of TBLASTN searches (data not shown). Genes in nematodes that are not one-to-one orthologs to C. elegans are named by subfamily and numbered and prefixed by br| and rm| (C. briggsae and C. remanei, respectively). #, a small-sized ABCA member unique in C. elegans; el|, C. elegans; dp|, D. pseudoobscura; dm|, D. melanogaster; mm|, Mus musculus; hs|, H. sapiens.
The presence of extensive orthology among nematode ABC proteins is in drastic contrast to what was observed between ABC genes of C. elegans and more distantly related organisms. For example, only 8 of 49 (16%) human ABC genes were found in one-to-one orthologous relationships with those of C. elegans, while an almost identical ABC gene set was found between mice and humans (Dean et al. 2001; Sheps et al. 2004). Rare orthology was also observed in a comparison between C. elegans and D. melanogaster or Saccharomyces cerevisiae. It is interesting to note that, despite poor orthology across these species, the overall numbers of ABC genes are roughly the same (Table 2). Widespread orthology within the three nematode species and within mammals and insects suggests that ABC family radiation took place concurrently with the radiation of the invertebrate phyla. Alternatively, the observed patterns were caused by ongoing lineage-specific birth–death of ABC genes. To measure gene rearrangements among the three nematode species, we examined inversion and deletion events around orthologous ABC genes. Six and four inversion events were observed involving C. briggsae and C. remanei ABC genes, respectively, in relation to those of C. elegans. Frequent deletion events were also observed (Table 1).
TABLE 2.
Subfamily distribution of ABC transporters in mammals, insects, and nematodes
| ABCA | ABCB | ABCC | ABCD | ABCE | ABCF | ABCG | ABCH | Total | |
|---|---|---|---|---|---|---|---|---|---|
| H. sapiensa | 12 | 11 | 12 | 4 | 1 | 3 | 5 | 0 | 48 |
| M. musculusa | 15 | 12 | 11 | 4 | 2b | 4 | 6 | 0 | 53 |
| D. melanogastera | 19 | 10 | 12 | 2 | 1 | 3 | 15 | 3 | 56 |
| C. elegans | 7 | 25 | 9 | 5 | 1 | 3 | 9 | 2 | 61 |
| C. briggsae | 6 | 23 | 9 | 5 | 1 | 3 | 9 | 2 | 58 |
| C. remanei | 7 | 23 | 9 | 5 | 1 | 3 | 9 | 2 | 59 |
This study.
Tandem duplications of ABC genes in nematodes:
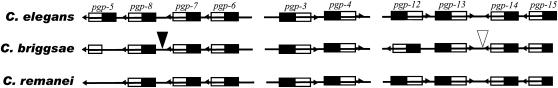
There are 16 tandemly duplicated ABC genes in the C. elegans genome, forming two four-gene clusters and four two-gene clusters (supplemental Figure 2 at http://www.genetics.org/supplemental/). Ten of them (the two four-gene clusters and one two-gene cluster) are composed of full-sized ABCB genes on the X chromosome (Figure 2). It is the duplication of these ABCB members that explains the expansion of subfamily B within nematode species (>22 members each) compared to other organisms (∼10 members each) (Table 2; supplemental Figure 1a at http://www.genetics.org/supplemental/). The three other two-gene clusters contain members of subfamilies C, D, and H (supplemental Figure 2 at http://www.genetics.org/supplemental/; Table 1). The tandemly duplicated ABC genes are well conserved among the three nematode species except for the set of genes consisting of C05A9.1, T21E8.3, -8.2, and -8.1, as described below. The presence of most of these tandemly duplicated ABC genes within the three species indicates that they were duplicated before the first speciation, which took place ∼80 million years ago (Stein et al. 2003). The four C. elegans genes, C05A9.1, T21E8.3, -8.2, and -8.1, form a clade, but instead of forming one-to-one orthologous relationships with their closest relatives in both C. briggsae and C. remanei, they appear to have arisen from species-specific duplications to form a cluster of paralogs, none of which can be unambiguously assigned one-to-one orthologs in the other two species even though they are present in synteny (Table 1; supplemental Figure 1a at http://www.genetics.org/supplemental/). There is also an ABC transporter pseudogene syntenic with C05A9.1 in C. briggsae while no such a fragment was found in C. remanei (Figure 2). It is interesting to note that a transposase-like and a reverse-transcriptase-like ORF are present in the two C. briggsae four-gene clusters, respectively. There is a lack of concordance among the inferred phylogenies for the four genes of the F22E10 cluster (supplemental Figure 1a at http://www.genetics.org/supplemental/), along with an apparent inversion of br|F22E10.1 in relation to F22E10.1 (Figure 2). The presence of a transposase-like ORF here points to a possible explanation for the complex history of this cluster.
Figure 2.—
Schematic of tandemly duplicated full-sized subfamily B genes present in syntenic regions in the three nematode species. (Top, middle, and bottom) Gene clusters in C. elegans, C. briggsae, and C. remanei, respectively. In C. elegans, the genes on the left and the right form two four-gene clusters while the two in the middle form a single two-gene cluster. The C. elegans genes in the left cluster are not orthologous to those in C. briggsae and C. remanei in the phylogenetic tree. The clusters are arranged by their order on chromosome X. Solid and open boxes represent ABC domains in the amino terminus and carboxyl terminus, respectively. Gene orientation is indicated by arrows. Solid and open triangles denote a reverse transcriptase gene and a transposase-like gene identified in the middle of the two C. briggsae four-gene clusters, respectively. br|F22E10.1 is in reverse orientation as opposed to F22E10.1(pgp-12). A residual ABC domain is found at the end of the C. briggsae syntenic region, which could be a remnant pseudogene (on the left).
A pairwise alignment of the syntenic genomic regions containing F22E10.1, 2, 3, and 4 between C. elegans and C. briggsae indicates excellent conservation of exon/intron structure (supplemental Figure 3 at http://www.genetics.org/supplemental/), suggesting that the cluster was formed before the divergence of the three nematode species. The alignment also highlights putative regulatory DNA elements within both 5′ and 3′ untranslated regions. Some of these conserved elements have been shown to be involved in the regulation of tissue-specific expression such as excretory cell-specific expression (Zhao et al. 2005; supplemental Table 1 at http://www.genetics.org/supplemental/).
Different subfamilies show differential divergence across species:
Despite the overall orthology of ABC genes among the three nematode species, different rates of amino acid divergence were often observed among different subfamilies. ABCA subfamily members show the least interspecies or intraspecies sequence similarity while those from ABCD, E, and F subfamilies demonstrate the highest conservation (Figure 1B; supplemental Figure 1, d and e, at http://www.genetics.org/supplemental/), suggesting that subfamily A has undergone faster divergence in amino acid sequence. Six ABCA members were identified in C. briggsae whereas seven were identified in both C. elegans and C. remanei. Only four of the seven C. elegans ABCA subfamily members form one-to-one ortholog trios in the three species (Figure 1A). F56F4.6, F55G11.9, and Y53C10A.9 result from specific expansion in C. elegans and form a co-orthologous group with its counterparts, rm|ABCA.1, -2, and -3 in C. remanei. C. briggsae has one member, br|ABCA.1, which is co-orthologous with the above six genes, while maintaining one ancient paralog, br|ABCA.2, that does not belong to any co-orthologous group. It is interesting that F56F4.6 has an unusually small size in relation to other ABCA members (260 aa vs. >1500 aa) whereas the syntenic br|ABCA.1 and rm|ABCA.2 have typical sizes (Table 1). ABCA members are also subject to uneven expansion between the two Drosophila species and between humans and mice (Figure 1A).
In addition to the four-gene cluster consisting of full-sized ABCB members that are undergoing species-specific expansion, as mentioned above (Figure 2; Table 1; supplemental Figure 1a at http://www.genetics.org/supplemental/), some of the ABC genes are unique in one or two of the three nematode species. For example, Y57G11C.1 is a half-sized ABC gene of the ABCB subfamily that could be a pseudogene: there is no EST match (WS160), the loss-of-function mutant (VC15) appears to be wild type (supplemental Table 2 at http://www.genetics.org/supplemental/), and its putative promoter did not drive observable GFP expression (Zhao et al. 2004). Loss of function for many other single ABC genes does not show a specific phenotype (supplemental Table 2 at http://www.genetics.org/supplemental/), suggesting possible functional redundancies between paralogous ABC members.
Gene rearrangements were frequently seen among the three species. Six and four inversions were observed around ABC genes in C. briggsae and C. remanei relative to those of C. elegans, respectively. Deletions were also found to be involved in the evolution of the ABC family (Table 1). Deletion or inversion events may be underestimated because some of the ABC genes are located at the ends of contigs, which makes it impossible to evaluate rearrangement events. An example is the case for rm|C47A10.1, which is present at the end of contig 703. Another example is br|Y43F8C.12, which is split by two contigs with few overlapping sequences (supplemental Table 3 at http://www.genetics.org/supplemental/). It is worth noting that a few ABC orthologs in C. briggsae or C. remanei are not present in the syntenic regions (Table 1). It remains unknown whether those were derived from transposition, reciprocal translocation, or other chromosomal rearrangements.
C. briggsae and C. remanei ABC genes are more related to each other than to those of C. elegans:
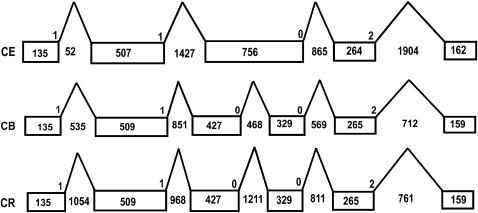
Comparisons among closely related species, as opposed to those among distantly related groups, are more likely to reveal evolutionary details that underlie life history. In addition, the time and the frequency at which evolutionary events such as intron gain or loss occurred may be obscured by comparison of distantly related species. Given the extensive conservation in the ABC family among nematode species, a comparison of ABC genes, especially those of the most conserved ones, could provide clues to the phylogeny among the three nematode species. Of the 53 ABC orthologous trios among the three nematode species, 39 C. briggsae and C. remanei ABC genes cluster with each other with the C. elegans ABC gene as an outgroup (Figure 1; supplemental Figure 1 at http://www.genetics.org/supplemental/), implying that C. briggsae and C. remanei are more related to each other than to C. elegans. This result is consistent with the recently resolved Caenorhabditis phylogeny using five different nuclear genes from 10 species (Kiontke et al. 2004) as well as other studies that used genes only from the same three species used here (Haag et al. 2002; Rudel and Kimble 2002). Further evidence came from comparison of the intron evolution of Y39E4B.1, an extremely conserved ABC gene and the single member of the ABCE subfamily. Sequences of ABCE proteins are highly conserved among all eukaryotic species, especially in their NBD domains, which show >90% identity across all eukaryotes (Kerr 2004). The br|Y39E4B.1 protein sequence has 98 and 95% identity to its C. remanei and C. elegans orthologs, respectively (data not shown). Notably, the ABCE subfamily has been found to have a single member in most eukaryotes, except in mice (Table 2; this study). The extreme conservation and one-to-one orthology makes this gene a promising candidate for inferring phylogenetic relations by comparison of its exon/intron evolution. To test the reliability of gene prediction for ABCE orthologs among the three nematode species, we performed an FGENESH ab initio gene prediction on the 40-kb syntenic regions, centering on the ABC coding sequence from all three species (see materials and methods). The predicted C. elegans exon/intron structure is identical to that annotated from EST evidence (WS160), indicating the high reliability of prediction of such a conserved ORF. The prediction results for br|Y39E4B.1 and rm|Y39E4B.1 show identical exon/intron structures, with six exons and five introns in the same locations and phases. In contrast, C. elegans Y39E4B.1 has five exons and four introns (Figure 3). Given relatively frequent intron loss within the nematode species (Kiontke et al. 2004), it is possible that Y39E4B.1 lost an intron in exon 3 because its size is equal to the combined sizes of exon 3 and 4 of br|Y39E4B.1 or rm|Y39E4B.1. In addition, the sizes of the remaining exons are also different between C. elegans and C. briggsae/C. remanei even though the three orthologs have the same protein sizes (Table 1). Taken together, these data suggest a closer relation between C. briggsae and C. remanei than between either of the two species and C. elegans, which is consistent with tree data from other conserved ABC genes.
Figure 3.—
Exon/intron structure of the single ABCE gene from the three nematode species. Syntenic genomic regions in C. briggsae and C. remanei centering on the Y39E4B.1 were used as input for FGENESH (Salamov and Solovyev 2000) using default parameters. The exon/intron size and positions were extracted from FGENESH output to draw the diagram. CE, Y39E4B.1; CB, br|Y39E4B.1; CR, rm|Y39E4B.1.
Positive selection in the ABC subfamilies:
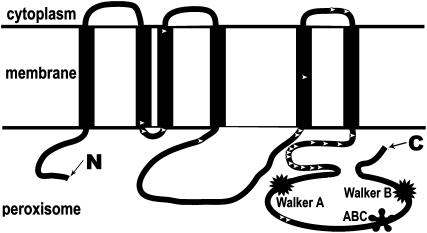
The presence and total size of the ABC gene family is highly conserved among closely related genomes despite being subject to fluctuation in subfamily size. It has been pointed out that the ABC gene family does not seem uniformly subject to one or the other mode of evolution (Sheps et al. 2004). For example, subfamilies E and F, which are not involved in membrane transport, have high rates of orthology over distantly related genomes, whereas subfamilies involved in transport are subject to faster evolution. We previously described that tandem-duplicated ABC genes seem subject to subfunctionalization. To examine whether there are any other mechanisms that are involved in shaping the ABC family dynamics, we examined the possibility of positive selection operating in the family. We began by measuring the nonsynonymous and synonymous substitution ratio (dN/dS) within individual C. elegans transport subfamilies because sequences within the same subfamily are usually similar in size and diverge from each other at reasonable distance. One exception is subfamily B, within which both full-sized and half-sized ABC proteins exist, as mentioned above. We treated these two groups as discrete data sets as described below. In each case, dN/dS was estimated by the ML approach, as implemented in the CODEML program (Yang 2000). Three pairs of the nested CODEML model were run on seven data sets consisting of ABCA, full-sized ABCB, half-sized ABCB, ABCC, ABCD, ABCF, and ABCG subfamilies, respectively (supplemental Table 4 at http://www.genetics.org/supplemental/). Strong evidence for positive selection was detected only in subfamily D (Figure 4; supplemental Figure 4 at http://www.genetics.org/supplemental/). To get a better understanding of how positive selection might affect protein function, we used the Bayes Empirical Bayes method of CODEML to determine specific sites with posterior probabilities of positive selection >0.85. These sites were mapped onto the secondary structure of a representative protein. ABCD subfamily members encode half transporters, which form hetro- or homo- dimers in vivo. All those that have been functionally characterized are expressed exclusively in the membrane of the peroxisome, where they regulate the transport of very-long-chain fatty acids (VLCFA) (Hettema et al. 1999). Strong positive selection sites were observed mostly within the region connecting the ABC domain and its adjacent transmembrane (TM) domain and less frequently in the three TM domains and intra- or extraperoxisomal regions. Two sites were found between the Walker A motif and the ABC signature motif, which form the highly conserved ABC domain together with the Walker B motif (Figure 4). Weak positive selection sites were also detected in subfamily A (data not shown). No significant positive selection was detected for the remaining data sets. A complete list of the sites under positive selection (BEB probability >0.85) is provided in supplemental Figure 4 at http://www.genetics.org/supplemental/. We did not test for positive selection for subfamilies E and H because the number of genes was too small to yield a confident estimation.
Figure 4.—
Distribution of positive selection sites (BEB probability >0.85) mapped onto the secondary structure of ABCD proteins. The ABCD gene C44B7.9 was used as the reference ABCD (see materials and methods). The ABC and TM domains were predicted by Pfam (Bateman et al. 2002) and TMHMM (Krogh et al. 2001), respectively. The open arrowheads denote positive selection sites. N, amino terminus; C, carboxyl terminus; ABC, ABC signature motif.
DISCUSSION
ABC transporters belong to one of the largest protein families that either import or export an unusually large spectrum of different substrates. In eukaryotes, ABC transporter genes are involved only in export (Saurin et al. 1999). Here we conducted a comparative analysis of the ABC family between the three closely related nematode species C. elegans, C. briggsae, and C. remanei and inferred the phylogenetic relationship among the genes by ortholog clustering in a phylogenetic tree and intron/exon structure comparison of ABCE members. Positive selection among the members of individual subfamilies was also examined.
Widespread orthology of ABC genes between nematode species:
ABC genes show frequent one-to-one orthology among C. elegans, C. briggsae, and C. remanei. In general, the orthologous trios formed well-separated groups on the phylogenetic tree, indicating that they have a common ancestor (Figure 1; supplemental Figure 1 at http://www.genetics.org/supplemental/). Frequent one-to-one orthology was also found between other groups of closely related species: humans and mice and the two Drosophila species. However, one-to-one orthology of ABC genes is poor when such a comparison is made between distantly related organisms. Good conservation among only closely related species implies that there are ongoing ABC gene birth-and-death events that take place in a lineage-specific manner.
The evolutionary pattern fits the model called “dynamic coherence,” which argues that gene families behave in a coherent fashion within the genome, i.e., that probabilities of duplication and deletion of genes within a gene family are not independent of each other (Huynen and van Nimwegen 1998). In agreement with this model, ABC gene subfamilies have undergone lineage-specific expansions or deletions while maintaining a similar number of genes in the family as a whole. For example, 10 tandemly duplicated ABC genes are found within subfamily B in C. elegans, which explains the expansion of the subfamily relative to non-nematode species (Table 2). However, nematode species have a smaller subfamily A than other organisms, which could be a consequence of gene deletion. Given that both the human and C. elegans full ABCB genes are involved in drug/toxin resistances (Broeks et al. 1996; Ling 1997), nematode-specific expansion of ABCB subfamily B possibly provides more efficient protection against toxins or other noxious materials present in their ecological niche, i.e., soil. Tandem duplications of ABC genes are not uncommon. For example, tandem duplications account for the lineage-specific expansions of subfamily A in mammals and subfamily G in insects, respectively (Dean et al. 2001). The eye-color genes white (w), brown (bw), and scarlet (st) of D. melanogaster encode three proteins that belong to subfamily G and are involved in the transport of guanine and tryptophan (precursors of the red and brown eye pigments) (Ewart et al. 1994). Nematodes have evolved a set of lineage-specific subfamily G members, of which the functions remain unknown (supplemental Figure 1f at http://www.genetics.org/supplemental/). Also, nematodes do not have ABC genes thought to be involved in cholesterol transport, which are shared by mammals and insects (Dean et al. 2001).
Investigation of both paralogous and orthologous members of large gene families can provide insight into genome evolution and phylogenetic relationships. It is interesting that most of the C. remanei ABC genes phylogenetically cluster with C. briggsae ones with those of C. elegans as its outside group. In agreement with this, exon/intron structures of the highly conserved ABCE proteins are more similar to each other between C. remanei and C. briggse than between either of the species and C. elegans, suggesting a closer relationship between C. briggsae and C. remanei than between either of the species and C. elegans. This agrees well with what has been reported (Kiontke et al. 2004), suggesting that the evolution pattern of ABC genes can provide useful insight for phylogeny.
Divergence of ABC genes between nematode species:
Despite frequent one-to-one orthology among the three nematode species, varying patterns of divergence among ABC genes are apparent. For example, ABCA subfamily members have diverged faster in amino acid sequence, on the basis of both the phylogenetic tree and the syntenic data, and three of the seven C. elegans ABCA members are not orthologous to single ABC genes in either C. briggsae or C. remanei. C. briggsae-specific clustering was not seen in the subfamily but two members, br|ABCA.1 and -2, still share an ancient ancestor with the above clusters (Figure 1A). The next closest relative of the three genes in C. elegans is C48B4.4/ced-7, which has been reported to be involved in engulfment of the cell corpses left behind by apoptosis (Wu and Horvitz 1998). Whether such an evolutionary pattern reflects differences in cell-corpse engulfment among nematode species remains to be explored.
Species-specific clustering of ABC genes was also observed in subfamily B members that resulted from tandem duplications. It is intriguing that four C. elegans-duplicated ABCB members, C05A9.1, T21E8.1, -2, and -3, are not orthologous to any gene in C. briggsae and C. remanei but a group of genes in the latter two species are nevertheless syntenic to the four C. elegans genes (Figure 2; Table 1), suggesting that the syntenic information may not necessarily reflect the origin of related genes. The presence of a transposase-like and reverse-transcriptase-like ORF within the two four-gene clusters suggests that transposition may have played a role in formation of this tandem cluster. It is increasingly clear that eukaryotic gene order is not random (Hurst et al. 2004). About half of the membrane protein genes in more than a dozen of sequenced genomes were found to be in tandem clusters (Kihara and Kanehisa 2000). Nematode genes appear to have a faster rate of rearrangement compared to those of Drosophila. We estimated that there is one rearrangement event for every five ABC genes between C. elegans and C. briggsae, which is approximately the same rate as previously reported, i.e., 4030 rearrangements for the whole genome between the two worms (Coghlan and Wolfe 2002).
The human ABCB subfamily contains a well-studied member, ABCB1/MDR1/PGP-1, which is involved in multiple drug resistance. The PGP1 protein is a broad-spectrum multidrug efflux pump with the common ABC protein structure of 12 transmembrane regions and two ATP-binding sites (Chen et al. 1986). The transmembrane regions are believed to bind hydrophobic drug substrates that are either neutral or positively charged (Ambudkar et al. 1999). C. elegans pgp genes (encoding full-sized ABCB members) have also been reported to be involved in drug resistance. For example, a pgp-3 (ZK455.7) mutant shows increased sensitivity to chloroquine and colchicines as well as to toxins produced by Pseudomonas auruginosa (Mahajan-Miklos et al. 1999). Both pgp-1 (K08E7.9) and pgp-3 mutants have been reported to be hypersensitive to the heavy metals cadmium and arsenate (Broeks et al. 1996). Expansion of the nematode B subfamily by duplications could play a major role in dealing with these environmental challenges.
Positive selection in ABC family:
Positive selection can be inferred from estimating rates of synonymous and nonsynonymous nucleotide substitution among the related genes. Strong evidence for positive selection at multiple sites was detected in subfamily D (Figure 4) and weaker evidence was found in subfamily A (data not shown).
The human ABCD1/ALDP gene encodes a half transporter, mutation of which caused adrenoleukodystrophy (Mosser et al. 1993). The gene is believed to be involved in the transport of VLCFAs across peroxisome membranes. All the human ABCD subfamily members identified so far are peroxisome exclusive half ABC transporter genes but their transcript abundance varies with tissues (Lombard-Platet et al. 1996). The ABCD genes are expected to be functional only after formation of homo- or hetrodimers. Homodimerization of ALDPs and heterodimerization of ALDP with ALDR or PMP70 have been demonstrated in vitro (Liu et al. 1999; Smith et al. 1999). Instead of four members, there are five ABCD subfamily members in C. elegans, two of which result from tandem duplication (Table 1; supplemental Figure 2 at http://www.genetics.org/supplemental/). Given the good orthology of the subfamily between C. elegans and Homo sapiens, it is likely that worm ABCD members play similar roles to those of mammals after forming a dimer. It is surprising that positive selection is observed within such a highly conserved subfamily. We speculate that positive selection sites within the ABCD subfamily could reflect the dynamic changes in food availability. C. elegans is a soil-dwelling nematode species that feed on bacteria. The spectrum of bacterial species varies with time and location. As a result, the nematode may experience changes in the source of VLCFAs, the substrates for a functional transporter formed by ABCD proteins. These changes could drive the observed positive selection.
Acknowledgments
We thank The Genome Sequencing Center at Washington University in Saint Louis and the Sanger Centre for permission to use the C. briggsae and C. remanei sequence. And we thank Andy Fire for the GFP vector and Ann Rose for dpy-5 mutants and rescue plasmid DNA. All the knockout mutant strains were generated by the C. elegans Knockout Consortium, which is funded by Genome Canada. Several strains were provided by the C. elegans Genetic Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by the Natural Sciences and Engineering Research Council of Canada, Genome British Columbia, and Genome Canada.
1Present address: Department of Genome Sciences, University of Washington, Seattle, WA 98195.
References
- Ambudkar, S. V., S. Dey, C. A. Hrycyna, M. Ramachandra, I. Pastan et al., 1999. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 39: 361–398. [DOI] [PubMed] [Google Scholar]
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller et al., 2002. The Pfam protein family database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., M. M. Clamp and R. Durbin, 2004. GeneWise and genomewise. Genome Res. 14: 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks, A., B. Gerrard, R. Allikmets, M. Dean and R. H. A. Plasterk, 1996. Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J. 15: 6132–6143. [PMC free article] [PubMed] [Google Scholar]
- Burge, C. B., and S. Karlin, 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Chen, C. J., J. E. Chin, K. Ueda, D. P. Clark, I. Pastan et al., 1986. Internal duplication and homology with bacterial transport proteins in Mdr-1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 47: 371–380. [DOI] [PubMed] [Google Scholar]
- Chen, N., S. Pai, Z. Zhao, A. Mah, R. Newbury et al., 2005. Identification of a nematode chemosensory gene family. Proc. Natl. Acad. Sci. USA 102: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A., and K. H. Wolfe, 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, M., A. Rzhetsky and R. Allikmets, 2001. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11: 1156–1166. [DOI] [PubMed] [Google Scholar]
- Ewart, D., D. Cannell, G. B. Cox and A. J. Howells, 1994. Mutational analysis of the traffic ATPase (ABC) transporters involved in uptake of eye pigment precursors in Drosophila melanogaster. Implications for structure-function relationships. J. Biol. Chem. 269: 10370–10377. [PubMed] [Google Scholar]
- Fitch, W., 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19: 99–113. [PubMed] [Google Scholar]
- Guindon, S., and O. Gascuel, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Haag, E. S., S. Wang and J. Kimble, 2002. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr. Biol. 12: 2035–2041. [DOI] [PubMed] [Google Scholar]
- Hettema, E. H., B. Distel and H. F. Tabak, 1999. Import of proteins into peroxisomes. Biochim. Biophys. Acta 1451: 17–34. [DOI] [PubMed] [Google Scholar]
- Higgins, C. F., and P. M. Sharp, 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Hurst, L. D., C. Pal and M. J. Lercher, 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5: 299–310. [DOI] [PubMed] [Google Scholar]
- Huynen, M. A., and E. van Nimwegen, 1998. The frequency distribution of gene family sizes in complete genomes. Mol. Biol. Evol. 15: 583–589. [DOI] [PubMed] [Google Scholar]
- Hyde, S. C., P. Emsley, M. J. Hartshorn, M. M. Mimmack, U. Gileadi et al., 1990. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346: 362–365. [DOI] [PubMed] [Google Scholar]
- Kerr, I. D., 2004. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 315: 166–173. [DOI] [PubMed] [Google Scholar]
- Kihara, D., and M. Kanehisa, 2000. Tandem clusters of membrane proteins in complete genome sequences. Genome Res. 10: 731–743. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101: 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., B. Larsson, G. von Heijne and E. L. Sonnhammer, 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Ling, V., 1997. Multidrug resistance: molecular mechanisms and clinical relevance. J. Cancer Chemother. Pharmacol. 40(Suppl.): S3–S8. [DOI] [PubMed] [Google Scholar]
- Liu, L., K. Janvier, V. Berteaux-Lecellier, N. Cartier, R. Benarous et al., 1999. Homo- and heterodimerization of peroxisomal ATP-binding cassette half-transporters. J. Biol. Chem. 274: 32728–32743. [DOI] [PubMed] [Google Scholar]
- Lombard-Platet, G., S. Savary, C. O. Sarde, J. L. Mandel and G. Chimini, 1996. A close relative of the adrenoleukodystrophy (ALD) gene codes for a peroxisomal protein with a specific expression pattern. Proc. Natl. Acad. Sci. USA 93: 1265–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, R., V. Silventoinen, S. Robinson, A. Kibria and W. Gish, 2003. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 31: 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos, S., M. W. Tan, L. G. Rahme and F. M. Ausubel, 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas auruginosa-Caenorhabditis elegans pathogenesis model. Cell 96: 47–56. [DOI] [PubMed] [Google Scholar]
- Mosser, J., A. M. Douar, C. O. Sarde, P. Kioschis, R. Feil et al., 1993. Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature 361: 726–730. [DOI] [PubMed] [Google Scholar]
- Nicholas, K. B., and H. B. J. Nicholas, 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments. Mol. Biol. Evol. 4: 406–425. [Google Scholar]
- Nielsen, R., and Z. Yang, 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel, D., and J. Kimble, 2002. Evolution of discrete Notch-like receptors from a distant gene duplication in Caenorhabditis. Evol. Dev. 4: 319–333. [DOI] [PubMed] [Google Scholar]
- Salamov, A. A., and V. V. Solovyev, 2000. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin, W., M. Hofnung and E. Dassa, 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48: 22–41. [DOI] [PubMed] [Google Scholar]
- Sheps, J. A., S. Ralph, Z. Zhao, D. L. Baillie and V. Ling, 2004. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 5: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, G. S., and E. Birney, 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 15: 6–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K. D., S. Kemp, L. T. Braiterman, J. F. Lu, H. M. Wei et al., 1999. X-linked adrenoleukodystrophy: genes, mutations and phenotypes. Neurochem. Res. 24: 511–525. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E. L., and E. V. Koonin, 2002. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 18: 619–620. [DOI] [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker, C., J. A. Sheps and A. M. Rose, 2006. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell. Mol. Life Sci. 63: 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. H., J. L. Kelley, H. M. Robertson, K. Ly and W. J. Swanson, 2005. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae. Proc. Natl. Acad. Sci. USA 102: 4476–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., and H. R. Horvitz, 1998. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93: 951–960. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2000. Phylogenetic Analysis by Maximum Likelihood (PAML), Version 3.0. University College London, London.
- Yang, Z., R. Nielsen, N. Goldman and A. M. Pedersen, 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., J. A. Sheps, V. Ling, L. L. Fang and D. L. Baillie, 2004. Expression analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J. Mol. Biol. 344: 409–17. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., L. Fang, N. Chen, R. C. Johnsen, L. Stein et al., 2005. Distinct regulatory elements mediate similar expression patterns in the excretory cell of Caenorhabditis elegans. J. Biol. Chem. 280: 38787–38794. [DOI] [PubMed] [Google Scholar]