Abstract
The maize transposable element Activator (Ac) has been exploited as an insertional mutagen to disrupt, clone, and characterize genes in a number of plant species. To develop an Ac-based mutagenesis platform for maize, a large-scale mutagenesis was conducted targeting the pink scutellum1 locus. We selected 1092 Ac transposition events from a closely linked donor Ac, resulting in the recovery of 17 novel ps1 alleles. Multiple phenotypic classes were identified corresponding to Ac insertions in the 5′-UTR and coding region of the predicted Ps1 gene. To generate a stable allelic series, we employed genetic screens and identified 83 germinally heritable ps1 excision alleles. Molecular characterization of these excision alleles revealed a position-dependent bias in excision allele frequencies and the predominance of 7- and 8-bp footprint products. In total, 19 unique ps1 excision alleles were generated in this study, including several that resulted in weak mutant phenotypes. The analysis of footprint alleles suggests a model of Ac excision in maize that is consistent with recent in vitro studies of hAT element excision. Importantly, the genetic and molecular methods developed in this study can be extended to generate novel allelic variation at any Ac-tagged gene in the genome.
THE maize transposable elements Activator (Ac) and Dissociation (Ds) are powerful insertional mutagens in plants and have been used to isolate genes (Kunze and Weil 2002), to define promoter and enhancer elements (Athma et al. 1992; Moreno et al. 1992; Sundaresan et al. 1995; Chin et al. 1999; Greco et al. 2003; Wu et al. 2003; Jin et al. 2004), and to create genetic mosaics for clonal analysis (Emerson 1917; Dawe and Freeling 1990; Peng and Harberd 1997; Jenik and Irish 2001). An important feature of both Ac and the nonautonomous derivative Ds is the tendency for short-range transposition. In studies of Ac transposition from the p1 and bz1 loci, nearly 60% of all transpositions were to sites within 5 cM of the donor Ac (dAc) (Greenblatt 1984; Dooner and Belachew 1989). This general feature of Ac/Ds has been exploited in maize to mutagenize genes that lie in close proximity to the donor element (Orton and Brink 1966; Brink and Williams 1973; Kermicle et al. 1989; Athma et al. 1992; Moreno et al. 1992; Weil et al. 1992; Alleman and Kermicle 1993).
The power of Ac as an insertional mutagen was perhaps most fully exploited in the dissection of the p1 locus. Utilizing Ac donor elements closely linked to p1, >400 Ac-induced p1 alleles have been generated (Orton and Brink 1966; Athma et al. 1992; Moreno et al. 1992). By carefully examining the variegation patterns of these alleles, promoter elements, enhancer elements, intron/exon boundaries, and critical amino acids were defined throughout coding and noncoding regions of the locus (Moreno et al. 1992). The elegance of such studies is perhaps most striking for maize, where time and cost constraints associated with constructing an allelic series of near-isogenic, stably expressed transgenic lines are prohibitive.
Ac and Ds can also be utilized to create genetically stable excision or “footprint” alleles. When Ac or Ds inserts into the genome, it generates an 8-bp target-site duplication (Muller-Neumann et al. 1984; Pohlman et al. 1984). Ac and Ds excision events are often imprecise (Baran et al. 1992; Moreno et al. 1992), resulting in the creation of novel footprint alleles (Muller-Neumann et al. 1984; Pohlman et al. 1984). At the waxy1 locus, two Ds excision alleles resulting in 9- and 6-bp insertions altered the enzymatic activity of the WAXY1 protein (Wessler et al. 1986). Similarly, Ds excision alleles at the r1 locus resulted in subtle perturbations of R function and defined critical domains in the R protein necessary for nuclear localization and protein–protein interactions (Liu et al. 1996, 1998). Interestingly, a Ds excision allele at shrunken2 resulted in an 11–18% increase in seed weight through the alteration of allosteric properties of the protein (Giroux et al. 1996). These studies have shown that the activity, localization, and function of a protein can be dramatically or subtly altered through Ac or Ds excision.
Despite the many useful features of Ac/Ds, relatively few genes have been characterized using these elements in maize. The primary limitation has been the lack of precisely mapped and evenly distributed Ac elements throughout the maize genome (Brutnell and Conrad 2003). However, in recent years several large-scale genomics programs have been developed to position Ac elements on genetic and physical maps of the maize genome (Cowperthwaite et al. 2002; Kolkman et al. 2005). The genetic materials created through these programs now provide the opportunity to exploit Ac to characterize thousands of genes throughout the maize genome.
To explore the potential of Ac as a tool in large-scale insertional mutagenesis programs, we conducted a regional mutagenesis of the pink scutellum1 (ps1) locus. The Ps1 gene encodes the enzyme lycopene β-cyclase that catalyzes the first committed step in xanthophyll biosynthesis (Singh et al. 2003). We previously utilized an Ac element located 4 cM from ps1 to clone the gene and generated seven unstable ps1 alleles (Singh et al. 2003).
In this study, we have selected 1092 independent Ac transposition events from the same donor Ac and recovered 17 novel ps1 alleles. Of the 17 alleles, 14 carried Ac insertions that were precisely positioned at the ps1 locus, whereas 3 of the ps1 alleles retained a signature of Ac excision. Thus, all of the recovered ps1 alleles were likely induced by Ac, but 18% did not carry a molecular tag for cloning. We have also developed a genetic strategy to enrich for the recovery of Ac footprint alleles. Sequence analysis of 57 footprint alleles was consistent with the “endonuclease model” of excision repair (Coen et al. 1989) whereby double-strand breaks are generated on each side of Ac following excision, resulting in the formation of hairpins at the donor locus. We propose that these hairpins are resolved by a selective endonucleolytic attack at a site immediately adjacent to the ligation site and discuss the implications of these findings for developing Ac/Ds-based mutagenesis programs in maize.
MATERIALS AND METHODS
Genetic stocks and selection schemes:
All alleles were maintained in the common genetic background of a color-converted W22 inbred line (Dooner and Kermicle 1971). Ac regional mutagenesis at ps1 from donor site bti97156∷Ac was previously described (Singh et al. 2003).
To identify rare Ac excision events, two genetic schemes were developed that exploit a Ds reporter to monitor Ac copy number. Ac activity was monitored with the Ds reporter, r-sc:m3, as previously described (Brutnell and Dellaporta 1994). In the absence of Ac, the Ds insertion at r-sc:m3 is stable, resulting in a colorless aleurone. In the presence of Ac, Ds excises at a time and frequency that is dependent on Ac copy number, resulting in purple sectored aleurone. An increase in Ac copy number delays Ds excision, resulting in kernels with smaller spots, and is generally referred to as the “negative dosage effect” (McClintock 1951).
In the first genetic scheme, kernels were selected from self-pollinated ears of lines carrying the donor Ac (bti97156∷Ac, dAc) and the ps1∷Ac allele (see supplemental Figure S1 at http://www.genetics.org/supplemental/). To enrich for Ac excision events, coarsely spotted kernels were selected, indicating that a single active Ac element was inherited through the male gametophyte. Because the donor and ps1 Ac insertions are closely linked in cis, recombination events are rare. To identify potential excision alleles, single-copy Ac kernels (dAc +/+ + or + ps1∷Ac/+ +) from self-pollinated dAc ps1∷Ac/+ + ears were planted in the field. DNA was extracted from seedling tissues and DNA blot analysis was performed as previously described (Singh et al. 2003) to identify plants that no longer carried Ac at the ps1 locus. Plants carrying these putative ps1 footprint alleles were self-pollinated, and ears were screened to identify families that segregated a single active Ac element and a ps1 mutant phenotype. Colorless kernels were selected from these ears (+ ps1/+ + or + +/+ +), and the plants self-pollinated to generate a stock that did not carry Ac.
A second genetic scheme was developed to identify novel ps1 alleles that condition a subtle mutant phenotype as described in the results. In this strategy, kernel selections were based solely on Ac copy number and not on ps1 mutant phenotypes. To select Ac excision alleles, lines doubly hemizygous for the dAc and the ps1∷Ac allele (dAc ps1∷Ac/+ +) were testcrossed to the r-sc:m3 tester line. Lines carrying a single Ac element were selected on the basis of kernel variegation pattern, and DNA was isolated from seedling tissues. To screen for the presence of the dAc, primers 53035.7 and TBP35 were used to amplify the DNA flanking the dAc insertion. Primer pairs used to amplify the Ac junction at the ps1 locus are listed in supplemental Table S1 and primer sequences are listed in supplemental Table S2 at http://www.genetics.org/supplemental/. Lines carrying putative ps1 excision alleles were self-pollinated to generate segregating families.
Fine mapping of ps1 alleles:
To precisely map Ac insertion sites at ps1, phenotypically wild-type kernels were selected from families segregating ps1 mutant kernels. DNA was extracted from seedling tissues and DNA blot analysis was performed to identify heterozygous individuals carrying an Ac insertion at ps1 using a ps1-specific DNA fragment (Singh et al. 2003). DNA flanking each of the Ac insertions and excision sites from ps1-d22, ps1-d23, and ps1-d24 were amplified using primers shown in Table S1 at http://www.genetics.org/supplemental/. To define ps1 footprint alleles, kernels carrying a single Ac were grown in the field and DNA was extracted from seedling tissue. PCR was performed to determine the presence of Ac at the donor site and ps1. Putative excision alleles were then amplified using Ps1-specific primers listed in Table S1 at http://www.genetics.org/supplemental/. As these primers amplified both footprint and wild-type Ps1 alleles, multiple PCR products were cloned and sequenced. The PCR reactions consisted of ∼30 ng DNA, 0.25 μm of each primer, 0.2 mm dNTP's (Promega, Madison, WI), 1× Taq polymerase buffer with MgCl2 (Promega), 0.5 m betaine, 4% DMSO, and 2 units of Taq polymerase (Promega). The reaction was denatured at 94° for 2 min and followed by 30 cycles of 94° for 30 sec, 57° for 45 sec, and 72° for 2 min. All cycles terminated with a final extension at 72° for 7 min. Amplification reactions were fractionated on 1.2% agarose gels and PCR products were purified using the QIAquick gel extraction kit (QIAGEN, Valencia, CA) following the manufacturer's recommendations. Products were ligated into pGEM T-easy vector (Promega) and sequenced as previously described (Singh et al. 2003).
Germination screen:
Maize kernels were sterilized in a 20% bleach solution by gentle rocking for 20 min followed by five rinses with sterile water and then was left to soak in water overnight at room temperature with gentle rocking. Ten kernels for each genotype were placed in a seed germination pouch (Mega International, St. Paul) and watered daily. Seed pouches were placed vertically in the growth chamber and grown under 40 μmol m2 s−1 white light. Germination rates were scored and plants were photographed 7 days after planting.
RESULTS
Insertional mutagenesis at Ps1 gene:
The Ps1 gene of maize was cloned utilizing the transposable element Ac as an insertional mutagen (Singh et al. 2003). In this previous study, seven independent Ac insertion alleles were recovered throughout the coding region of Ps1 from a population of 386 transposition events. To further characterize the genetic fine structure of the ps1 locus, we expanded the scope of the mutagenesis program. An additional 1092 transposition events were recovered as single kernel selections from an Ac donor (bti97156∷Ac) resident 4 cM from the ps1 locus. The resulting self-pollinated ears were screened and 17 families were identified that segregated pink/viviparous kernel phenotypes.
To characterize these putative ps1 alleles, DNA blot analysis was first performed to map potential Ac insertion sites (see materials and methods). This survey indicated that in 14 of 17 alleles, an Ac element was present at the ps1 locus. However, we did not detect an Ac element or any large-scale rearrangements of ps1 sequence associated with the remaining 3 ps1 alleles. Thus, we sequenced the 5′-UTR complete coding region and the 3′-UTR of the Ps1 gene from each of the 3 alleles. Comparison of the progenitor W22 Ps1 sequence with the sequence from the mutant alleles identified polymorphic sites in each of the non-Ac-tagged alleles. In each case, there was a small duplication of the ps1 sequence. Derivative ps1-d22 carried a 4-bp insertion, ps1-d23 carried an 8-bp insertion, and ps1-d24 carried an 8-bp insertion at positions 925, 1253, and 1857, respectively, of the Ps1 gene (GenBank AY206862). These insertions corresponded to tandem duplications of Ps1 sequence typical of Ac excision events (Scott et al. 1996). In summary, of the 1092 kernels selected as carrying a tr-Ac, 14 families were recovered with Ac insertions at ps1, and 3 ps1 mutant alleles that did not carry an Ac insertion were recovered. Thus, Ac-induced alleles of ps1 were recovered in 1.6% of kernel selections (17/1092), and 18% of the ps1 alleles recovered were not tagged with an Ac element.
DNA flanking each Ac insertion was amplified and sequenced to precisely position the Ac insertion within the ps1 locus (see materials and methods). Insertions that resulted in a mutant phenotype were identified in 5′-UTR and throughout coding regions as summarized in Figure 1A. Ac elements were distributed in both orientations and at roughly equal distribution throughout the ps1 locus (χ2 = 1.19, d.f. = 1, P > 0.2). A uniform distribution of Ac is consistent with studies of Ac insertion at p1 (Athma et al. 1992; Moreno et al. 1992) and transpositions of Ac from bz1 and waxy1 (Cowperthwaite et al. 2002) where no insertion site preference for the proximal or distal half of genes was detected. This is in marked contrast to the distribution of Mutator elements in maize or Ds insertions in Arabidopsis that preferentially target the 5′ half of genes (Parinov et al. 1999; May et al. 2003).
Figure 1.—
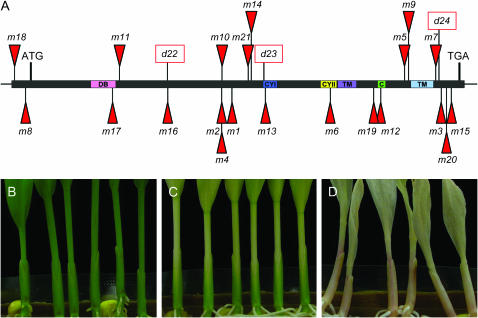
Ac-induced ps1 allelic variation. (A) Ac insertions in ps1. The long horizontal black bar represents the coding and noncoding regions of the intronless Ps1 gene. Red triangles above the gene represent Ac insertions in 5′–3′ orientation in each ps1-m∷Ac allele; red triangles below the gene are Ac insertions in 3′–5′ orientation. Orientation is related to Ac transcriptional start sites. Rectangular open boxes represent Ac-induced ps1-d alleles with the signature of Ac excision. Previously identified conserved regions (Cunningham et al. 1996) are shown by colored boxes: DB, dinucleotide-binding motif; CYI, cyclase motif I; CYII, cyclase motif II; TM, predicted transmembrane helix; C, charged region. Representative seedling phenotypes that are homozygous for (B) the W22 Ps1 allele, (C) ps1-m18∷Ac, and (D) ps1-m7∷Ac are shown.
Although the Ps1 gene contains no introns, Ac insertions at ps1 conditioned a range of phenotypic variation that was position dependent. To quantitate this variation, we exploited the fact that the Ps1 gene is required for both abscisic acid (ABA) production and normal carotenoid content of leaf tissues (Singh et al. 2003). A loss of ABA production leads to precocious germination due to a failure to suppress germination late in the grain-filling process. These kernels do not survive the desiccation process following harvest. Thus, the percentage of germination is an indirect measurement of ABA activity and PS1 function.
Seed germination assays were performed as described in materials and methods (data are in Table 1). Ac insertions in the middle of the Ps1 gene resulted in a severe mutant phenotype, characterized by vivipary and a failure to germinate after the ear was dried. These phenotypes are consistent with the absence of functional PS1 protein, thus preventing the accumulation of ABA. Insertions in the 5′-UTR and near the carboxy terminus of the predicted protein were less severe. For example, germination rates for seedlings homozygous for both the ps1-m8∷Ac and the ps1-m18∷Ac allele were 98%. In contrast, Ac insertions at the 3′-end of the Ps1 gene conditioned much lower germination rates. Individuals homozygous for ps1-m7∷Ac, ps1-m3∷Ac, ps1-m20∷Ac, and ps1-m15∷Ac conditioned germination rates of 88, 59, 5, and 34%, respectively. Thus, the site of Ac insertion in the Ps1 gene has a dramatic effect on germination rate.
TABLE 1.
Seed germination ratio of ps1∷Ac alleles
| ps1∷Ac allelea | Ac orientation | % germinationb | Seedling phenotype | Seedling lethal |
|---|---|---|---|---|
| Ps1-W22 | NA | 100 (100/100) | Green | No |
| ps1-m18∷Ac | 5–3′ | 98 (59/60) | Virescent | Yes |
| ps1-m8∷Ac | 3′–5′ | 98 (59/60) | Pale green mesocotyl | No |
| ps1-m17∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m11∷Ac | 5′–3′ | 0 (0/20) | NA | Yes |
| ps1-m16∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m2∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m4∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m10∷Ac | 5′–3′ | 0 (0/20) | NA | Yes |
| ps1-m1∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m21∷Ac | 5′–3′ | 0 (0/20) | NA | Yes |
| ps1-m14∷Ac | 5′–3′ | 0 (0/20) | NA | Yes |
| ps1-m13∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m6∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m19∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m12∷Ac | 3′–5′ | 0 (0/20) | NA | Yes |
| ps1-m5∷Ac | 5′–3′ | 0 (0/20) | NA | Yes |
| ps1-m9∷Ac | 5′–3′ | 0 (0/20) | NA | Yes |
| ps1-m7∷Ac | 5′–3′ | 88 (35/40) | White with green tip | Yes |
| ps1-m3∷Ac | 3′–5′ | 59 (53/90) | White with green tip | Yes |
| ps1-m20∷Ac | 3′–5′ | 5 (2/40) | White with green tip | Yes |
| ps1-m15∷Ac | 3′–5′ | 34 (20/59) | Virescent | Yes |
ps1∷Ac alleles are listed according to the Ac insertion site in the Ps1 gene.
Germination ratio was scored as the average of three replicates.
Insertions in the 5′-UTR permit the accumulation of ABA pools that are sufficient to protect the embryo during desiccation, leading to very high rates of germination. Insertions in the central and 3′ region of the gene are likely to produce truncated protein products that disrupt PS1 function, thereby limiting ABA production. The reduced levels of ABA may result in delayed developmental arrest and reduce the desiccation tolerance of the embryo, resulting in a greatly reduced germination rate.
In addition to ABA production, PS1 function is also required for the synthesis of photoprotective carotenoids. As observed for germination rates, the position of the Ac insertion at Ps1 also had a dramatic effect on the seedling phenotype (Figure 1, C and D). Insertions in the 5′-UTR, including ps1-m18∷Ac and ps1-m8∷Ac, resulted in seedlings with slightly pale green leaf sheath tissue and virescent leaf blade tissue (Figure 1C). In contrast, insertions near the 3′ region of the gene, including ps1-m7∷Ac, ps1-m3∷Ac, ps1-m15∷Ac, and ps1-m20∷Ac, resulted in albescent or slightly pink leaf tissues, due to the accumulation of lycopene in seedling tissues (Figure 1D; Singh et al. 2003). Thus, the leaf phenotypes correlated well with the germination assays, suggesting that Ps1 functions similarly in seed and seedling tissues.
Creating novel alleles of ps1 by Ac excision:
To fully exploit Ac as tool to create novel and genetically stable allelic variation, we developed two genetic schemes to rapidly identify germinally inherited Ac excision alleles on the basis of canonical “negative dosage effect” of Ac (McClintock 1951). The first scheme (see materials and methods) was dependent on identifying a ps1 mutant phenotype from self-pollinated ears of ps1∷Ac heterozygous plants (see supplemental Figure S1 at http://www.genetics.org/supplemental/). Thirty-seven putative ps1 alleles were generated and 11 ps1 excision alleles were further characterized using this selection method (Table 3, scheme I). One of the limitations of this scheme is the potential to miss subtle mutant phenotypes that are the most informative for structure–function studies as discussed below. Thus, we developed a second strategy that was used to recover the majority of the Ac excision alleles. This second strategy was based on a two-step screen that was solely dependent on detecting germinal Ac excision events and not ps1 phenotypic variation. Importantly, this scheme can be readily applied to any Ac insertion that is closely linked to a donor insertion and is illustrated in Figure 2A.
TABLE 3.
Novel ps1 footprint alleles
| ps1 allele | Footprint sequence | Size (bp) | No. of alleles | Schemea | Allele severityb | |
|---|---|---|---|---|---|---|
| ps1-m17∷Ac | GCGCCATC | GCGCCATC | Ac | Strong | ||
| GCGCCATg | cCGCCATC | 8 | 3 | II | Strong | |
| ps1-m2∷Ac | GGTACCAG | GGTACCAG | Ac | Strong | ||
| GGTACCAc | GTACCAG | 7 | 1 | I | Strong | |
| ps1-m21∷Ac | CCCCGAAG | CCCCGAAG | Ac | Strong | ||
| CCCCGAA | CCCGAAG | 6 | 1 | II | Wild type | |
| ps1-m14∷Ac | CCGAAGGG | CCGAAGGG | Ac | Strong | ||
| CCGAAGG | gCGAAGGG | 7 | 2 | II | Strong | |
| ps1-m13∷Ac | GCCATGCC | GCCATGCC | Ac | Strong | ||
| GCCATGCg | cCCATGCC | 8 | 2 | II | Strong | |
| GCCATGCgg | CCATGCC | 8 | 2 | II | Strong | |
| ps1-m6∷Ac | GGTGCACC | GGTGCACC | Ac | Strong | ||
| GGTGCAC | cGTGCACC | 7 | 3 | II | Strong | |
| GGTGCACg | GTGCACC | 7 | 3 | II | Strong | |
| GGTGCAC | GTGCACC | 6 | 1 | II | Strong | |
| ps1-m12∷Ac | GCGGCAGA | GCGGCAGA | Ac | Strong | ||
| GCGGCAG | cCGGCAGA | 7 | 17 | II | Strong | |
| GCGGCAGt | cCGGCAGA | 8 | 6 | II | Strong | |
| GCGGCAGt | CGGCAGA | 7 | 1 | II | Strong | |
| ps1-m9∷Ac | GCCACACT | GCCACACT | Ac | Strong | ||
| GCCACAC | ggcCCCACACT | 9 | 1 | I | Weak | |
| ps1-m7∷Ac | GTCGAGGC | GTCGAGGC | Ac | Strong | ||
| GTCGAGGg | cTCGAGGC | 8 | 6 | I | Strong | |
| GTCGAGG | cTCGAGGC | 7 | 1 | I | Strong | |
| ps1-m3∷Ac | AAGGGCAC | AAGGGCAC | Ac | Strong | ||
| AAGGGCAg | tAGGGCAC | 8 | 2 | I, II | Strong | |
| AAGGGCA | GGCAC | 4 | 1 | I | Strong | |
| ps1-m15∷Ac | TTGGCAAC | TTGGCAAC | Ac | Strong | ||
| TTGGCAA | aTGGCAAC | 7 | 2 | II | Strong | |
| TTGGCAA | TGGCAAC | 6 | 2 | II | Weak | |
Underlined sequences represent 8-bp duplications flanking the original Ac insertion.
Scheme (I or II) refers to the method used to identify the excision allele as detailed in materials and methods.
Allelic strength is based on germination assay: ≥90%, weak; <90%, strong.
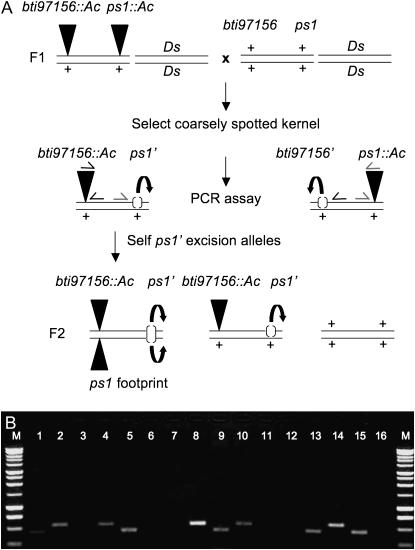
Figure 2.—
Genetic scheme II used to identify novel Ac excision alleles. (A) Genetic strategy developed to recover ps1 footprint alleles. Triangles represent Ac insertions. “+” represents the wild-type allele. Large parentheses represent putative excision alleles. Ac excision events are indicated by curved arrows. Solid arrows denote primers designed to amplify the donor Ac (dAc, bti97156∷Ac). Shaded angled arrows represent primers designed to amplify the Ac insertion site at ps1 locus. (B) A representative PCR gel image. When Ac is present, Ac-specific and locus-specific primers result in amplification products of a predicted size. If Ac is absent from the locus, no products are detected. The 1-kb DNA ladder (Promega) is labeled as M. Eight individual plants were genotyped using two PCR reactions. Odd-numbered lanes show products that result from amplification of the dAc (lanes 1, 5, 9, 13, and 15), whereas even-numbered lanes show expected products when Ac is inserted at the ps1 locus (lanes 2, 4, 8, 10, and 14). Lanes without amplification products indicate a potential Ac excision allele.
To generate transpositions using the two-step strategy, plants hemizygous for both the Ac donor (bti97156∷Ac) and the ps1∷Ac allele were testcrossed by pollen carrying the Ds reporter gene r-sc:m3 (see materials and methods). The majority of the testcross progeny were expected to carry both Ac alleles or no Ac as the two Ac insertions are tightly linked in cis (<4 cM). However, Ac excision events that result in the loss of Ac from the genome will generate chromosomes carrying a single Ac element. These kernels were identified as coarsely spotted due to the decrease in Ac copy number (see materials and methods). The second step was to discriminate between excision alleles from ps1 and the donor Ac loci using a PCR-based strategy. Two pairs of primers were designed to amplify the junction of the Ac and flanking DNA. If Ac excision occurred at the donor site, a PCR product was expected from the ps1 insertion site and not from the donor allele. Conversely, when excision occurs from the ps1 locus, only the Ac donor site is expected to amplify in the PCR assay. Once the putative ps1 footprint alleles were identified, PCR products spanning the excision sites were cloned and sequenced as described in materials and methods. Plants carrying putative ps1 excision alleles were then self-pollinated and progeny were screened for ps1 phenotypic variation.
Utilizing the second genetic scheme, we identified 342 coarsely spotted kernels from testcross ears as carrying putative Ac excision alleles. Seedlings were genotyped for the presence or absence of bti97156∷Ac and ps1∷Ac as described above. A representative gel image is shown in Figure 2B. Two PCR reactions were performed for each of the 342 DNA samples examined. As shown in Figure 2B (lanes 3, 4 and 7, 8), PCR products were detected when using primers specific for the Ac insertion at ps1, and no product was amplified using primers specific to the donor allele. These results indicated that the individuals retained the Ac insertion at ps1 and were not characterized further. Amplification products from putative ps1 excision alleles are also shown in Figure 2B (lanes 5, 6 and 15, 16). Here, the expected band sizes were detected using the donor-Ac primer pair, but not with the ps1-specific primers, suggesting that the closely linked dAc was present and that the Ac at ps1 had excised. These hemizygous individuals were grown to maturity and self-pollinated to generate families segregating a putative Ac excision allele.
Two exceptional classes were also detected. In 36 individuals, amplification products were detected with both sets of primers, indicating that both Ac insertions were still present. As discussed below, these events were likely recovered due to nonconcordance of embryo and endosperm genotypes. A second exceptional class of 163 seedlings did not contain either parental Ac allele. Our inability to detect the parental Ac insertions was not likely due to failed PCR reactions as positive controls were performed for both PCR reactions and DNA samples. Instead, it is likely that selections for a single copy of Ac in the genome led to the recovery of unlinked transpositions that were inherited with the non-Ac-containing parental chromosome. These results are summarized in Table 2 and show that, of the 342 kernel selections, 46 putative ps1 excision alleles were identified (13% of total) from eight independent ps1∷Ac alleles.
TABLE 2.
Ac excision frequency at different loci in ps1
| ps1 allele | Ac insertion site | ps1∷Ac excised | Donor Ac excised | Both Ac's present | Neither Ac present | Total |
|---|---|---|---|---|---|---|
| ps1-m18∷Ac | 5′-UTR | 0 | 1 | 0 | 1 | 2 |
| ps1-m8∷Ac | 5′-UTR | 0 | 17 | 7 | 8 | 32 |
| ps1-m17∷Ac | Dinucleotide-binding motif | 3 | 3 | 4 | 9 | 19 |
| ps1-m11∷Ac | 0 | 4 | 1 | 3 | 8 | |
| ps1-m10∷Ac | 0 | 1 | 1 | 6 | 8 | |
| ps1-m21∷Ac | 1 | 0 | 0 | 2 | 3 | |
| ps1-m14∷Ac | 2 | 2 | 1 | 7 | 12 | |
| ps1-m13∷Ac | Cyclase motif 1 | 4 | 1 | 2 | 17 | 24 |
| ps1-m6∷Ac | Cyclase motif 2 | 7 | 3 | 10 | 6 | 26 |
| ps1-m12∷Ac | Charged region | 24 | 41 | 9 | 20 | 94 |
| ps1-m3∷Ac | 3′-end | 1 | 13 | 0 | 40 | 54 |
| ps1-m20∷Ac | 3′-end | 0 | 1 | 0 | 1 | 2 |
| ps1-m15∷Ac | 3′-end | 4 | 10 | 1 | 43 | 58 |
| Total | 46 | 97 | 36 | 163 | 342 |
Novel stable ps1 footprint alleles:
In total, 83 stable ps1 alleles were generated from both selection schemes (37 from the first scheme and 46 from the second). However, only 19 unique ps1 excision alleles were generated (Table 3). The relatively small number of unique alleles is due to the predominance of one or two excision products associated with each Ac insertion. For example, of the 24 independent ps1 footprint alleles recovered from ps1-m12∷Ac, 17 carried an identical 7-bp insertion, 6 carried an identical 8-bp insertion, and 1 had a unique 7-bp insertion. Thus, among the 24 excision products generated from ps1-m12∷Ac, a 7-bp insertion is the predominant footprint sequence and only 3 unique footprint alleles were created. The majority of the excision events showed imperfect 7- or 8-bp duplications of ps1 sequence at the site of Ac insertion. In most cases, the nucleotides immediately flanking the Ac insertion suffered transversion mutations (e.g., C to G).
Phenotypic diversity at ps1 locus through stable allelic series:
Although most ps1 excision alleles retained a 7- or 8-bp imperfect target-site duplication, we identified four ps1 mutants that carried 6- or 9-bp insertions. As shown in Table 3, footprint allele ps1′-m9.1 carries a 9-bp insertion derived from ps1′-m9∷Ac. This allele is predicted to encode a novel PS1 protein carrying an in-frame three-amino-acid insertion at the Ac excision site. Footprint alleles ps1′-m6.1, ps1′-m21.1, and ps1′-m15.1 carry 6-bp insertions and are derived from ps1-m6∷Ac, ps1-m21∷Ac and ps1-m15∷Ac, respectively. Each insertion allele is predicted to encode a protein with an in-frame two-amino-acid insertion at the Ac excision site.
As shown in Figure 3B, individuals homozygous for these four footprint alleles display a range of phenotypic variation from near wild type to embryo lethality. Excision allele ps1′-m21.1 carries an insertion in a nonconserved region of the PS1 protein (Figure 3A). Seed and seedling tissues were phenotypcially indistinguishable from wild type, and germination was 100% in the seed germination assay. Excision allele ps1′-m15.1 also conditioned a 100% germination rate but seedlings were slightly pale green with pink mesocotyl tissue. The two-amino-acid insertion in ps1′-m15.1 is close to the 3′-end of the Ps1 gene but at a site that is conserved among Ps1 orthologs and suggests that even a slight perturbation of protein sequence at this site negatively affects protein stability or activity. Kernels homozygous for the ps1′-m9.1 allele were slightly pink and seeds germinated at a rate of 90%. Geminated seedlings were virescent and died at the third-to-fourth leaf stage. Although this mutant phenotype is more severe than either the ps1′-m21.1 or the ps1′-m15.1 allele, it is less severe than the parental ps1-m9∷Ac allele, suggesting that the excision allele encodes a PS1 protein with limited activity. The three-amino-acid insertion in ps1′-m9.1 is at a relative conserved region but closer to the central portion of the PS1 protein. Interestingly, the two-amino-acid insertion present in ps1′-m6.1 is located in the highly conserved cyclase motif II (Singh et al. 2003). This conserved motif has been hypothesized to play a role in generating β-ionone rings from lycopene that are necessary for the production of the ABA precursors γ-carotene and β-carotene (Cunningham et al. 1996). Seedlings homozygous for ps1′-m6.1 display a very strong ps1 mutant phenotype characterized by a pink endosperm, viviparous kernels, and a 0% germination rate. Given that this in-frame insertion is most severe, it is likely that this insertion disrupts the β-ring cyclization function of the enzyme, thus preventing production of γ- and β-carotenes and inducing the accumulation of lycopene. In the absence of xanthophylls, ABA also fails to accumulate, resulting in a viviparous phenotype.
Figure 3.—
ps1 footprint alleles carrying in-frame insertions. (A) Plant lycopene-β-cyclase partial sequence alignments (Singh et al. 2003). The sequence of in-frame amino acid insertions in each ps1 footprint allele is denoted by arrows. (B) Phenotypes of seedlings homozygous for ps1 alleles induced by in-frame amino acid insertions. Seedlings homozygous for the ps1′-m6.1 allele failed to germinate.
DISCUSSION
Ac regional mutagenesis:
In this study, we conducted a large-scale mutagenesis of the ps1 locus to examine the efficacy of Ac mutagenesis in maize. Using a closely linked Ac element as a donor locus, we identified 17 independent Ac-induced ps1 alleles. DNA blot and sequence analysis indicated that all but 3 ps1 alleles recovered were due to a single Ac insertion. The other 3 alleles carried a signature of Ac excision, indicating that all mutants recovered were induced by Ac. Nevertheless, 18% of recovered mutants did not carry an Ac insertion. In the context of Ac mutagenesis programs, these findings suggest that 15–20% of mutants recovered from such programs might not carry Ac insertions. Indeed, in a comprehensive survey of 1225 Ac transposition events generated from the waxy1 and bz1 loci, mutant phenotypes cosegregated with Ac in only 10% of the families (Cowperthwaite et al. 2002). Thus, it is likely that at least some of the mutants recovered were the result of Ac or Ds insertion and subsequent excision that generated nonfunctional alleles. However, it is likely that Ds and Ac excision alleles represent a minor proportion of mutants recovered in nondirected Ac-tagging programs. Assuming a similar rate of Ac footprint allele formation as observed here, at most 2% of the mutant families recovered in the Cowperthwaite study would be the result of Ac excision alleles (∼20% of the Ac-induced mutations). As previously discussed (Cowperthwaite et al. 2002), the increased rate of mutation associated with the presence of an active Ac element in the genome, may be the result of an increased rate of Ds element transposition. However, it is also possible that the presence of an active transposon in the genome increases the rate of spontaneous mutation or reduces the efficiency of DNA damage repair mechanisms.
A genetic scheme for generating footprint alleles:
Although Ac and Ds are clearly useful tools for site-directed mutagenesis, one serious drawback is that very large populations must be screened to identify rare excision events that lead to subtle alterations in protein structure and visible mutant phenotypes. Thus, it is perhaps not surprising that previous studies have been restricted to the analysis of genes that condition obvious seed phenotypes (e.g., waxy1, r1, bz1, and sh2) (Wessler et al. 1986; Schiefelbein et al. 1988; Giroux et al. 1996; Liu et al. 1996, 1998). To expand the repertoire of tools for the global analysis of gene function in maize, we have developed a high-throughput genetic screen to enrich for independent and germinally heritable Ac excision alleles. This screen is based on Ac/Ds-mediated variegation and thus is independent of phenotypic variation conditioned by the gene of interest.
Two genetic schemes were developed to identify novel stable ps1 footprint alleles that exploit the negative dosage of Ac to select for excision events. An essential feature of these approaches is that the two Ac elements are closely linked in cis. Otherwise, recombination between two loosely linked Ac insertions would result in a number of false positives. However, as Ac tends to insert at closely linked sites (Greenblatt 1984; Dooner and Belachew 1989), most schemes that have been developed to utilize Ac in directed tagging strategies exploit this tendency for closely linked transposition (Dellaporta and Moreno 1994; Brutnell and Conrad 2003). Thus, the majority of Ac-tagged mutants should carry a dAc closely linked in cis and be amenable to the strategies outlined here.
As discussed above, we favor the scheme in which Ac excision events were identified solely through changes in Ac copy number, thus enabling the recovery of weak ps1 alleles that may escape phenotypic screens. Indeed, one of the alleles generated in this study (ps1′-m21.1) was phenotypically indistinguishable from wild type yet carried a predicted two-amino-acid insertion in the coding region. Using this scheme, we attempted to generate germinal Ac excision alleles from ps1∷Ac insertions that were distributed throughout coding and noncoding regions of ps1 and recovered germinal excision alleles from eight alleles. Surprisingly, we did not detect Ac excision alleles from all of the ps1∷Ac alleles (Table 2). Furthermore, we observed an asymmetry in excision frequency between the donor Ac and several ps1 insertion alleles. Approximately two times as many alleles were recovered as a result of Ac excision from the donor Ac relative to the ps1 insertion (97:46, respectively, in Table 2). In particular, all Ac excision events recovered from lines carrying ps1-m8∷Ac were generated from the donor locus (17/17). It is unclear what may have generated this bias in excision allele frequency. It is possible that differences in methylation status at the Ac element itself may influence excision frequencies. Ac elements with increased methylation show greatly reduced rates of germinal excision (Brutnell and Dellaporta 1994; Brutnell et al. 1997). However, all Ac insertions examined at ps1 contributed to Ac dosage and thus were likely to be largely hypomethylated (Chomet 1988). A similar bias in excision frequency has been reported for Ds insertions at the waxy1 locus (Eisses et al. 1997). In this instance, a dominant suppressor of gametophytic Ds excision that was closely linked to the Ds insertion at waxy1 was identified. A closer examination of Ac methylation in ps1-m8∷Ac may help to clarify the likely mechanism underlying the low frequency of germinal excision events.
In addition to identifying Ac excision alleles, two exceptional kernel classes were also recovered. Of the 342 seedlings that were genotyped, 163 that did not carry either the dAc or the ps1∷Ac insertion were identified (Table 2). The majority of these seedlings likely carried Ac insertions unlinked to the ps1 locus that cosegregated with the non-Ac-containing parental chromosome. The other seedling class carried both parental Ac alleles. These progeny were likely the result of Ac excision events that occurred during a mitotic division of megagametogenesis. In this scenario, the megaspore mother cell inherits both parental Ac's. During a subsequent mitotic division, Ac excises from either the donor locus or ps1, reducing the copy number of Ac in the lineage fated to give rise to a polar cell. This excision event is not inherited by the lineage contributing to the egg nucleus, leading to a coarsely spotted endosperm and nonconcordance of endosperm and embryo genotypes. As 36 seedlings were found to carry both Ac insertions, we calculate a minimal frequency of nonconcordance at 11%. For comparison, ∼40% of all Ds excisions from the r1 locus mediated by Ac-im occur during gametophytic development (Conrad and Brutnell 2005) whereas at least 34% of Ac transpositions from the bz-m2(Ac) likely occur at a cell division during megagametogenesis (Dooner and Belachew 1989). Thus, our calculated frequency of nonconcordance is likely to be an underestimate of the true frequency of gametophytic transposition. Indeed, additional scenarios can be envisioned where excision of Ac during gametophytic development would lead to the recovery of kernels that lack both parental Ac elements in the embryo, but that had inherited a single copy of Ac in the endosperm lineage. Thus, some of the events scored as unlinked transposition events (163) are likely ps1 or dAc excision alleles. In summary, 342 kernel selections resulted in the recovery of 46 ps1 excision alleles from 8 ps1∷Ac insertions, and 13% (46/342) of the total kernel selections resulted in stable ps1 footprint alleles. Given that the rate of germinal Ac excision is ∼1–2% (Brutnell and Dellaporta 1994), our selection scheme results in an ∼10-fold enrichment for germinally heritable Ac excision alleles.
Mechanism of footprint formation:
One of the most comprehensive studies of Ds excision was conducted by Weil and colleagues at the waxy1 locus (Scott et al. 1996). In their study, 621 somatic transposition events were examined from six Ds insertions distributed throughout the waxy1 locus. They observed that (1) multiple excision products are generated from each Ds allele, (2) the predominant excision product varies with Ds insertion site, and (3) the frequency of each class of excision product varies for each insertion. Here, we have examined only Ac excision events that are germinally inherited and have likely recovered many that occurred during gametophytic development. Nevertheless, our findings are largely congruent with the findings of Weil and colleagues, suggesting that the mechanism of Ac excision and repair does not differ significantly from Ds excision across tissue types and developmental stages in maize.
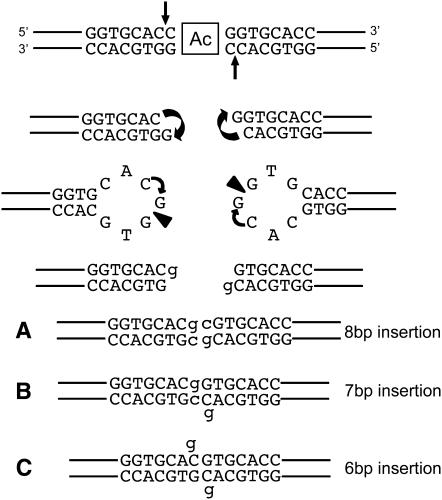
Studies of Ds excision in yeast (Weil and Kunze 2000) and Hermes excision in vitro (Zhou et al. 2004) strongly suggest a model of hAT element excision in which a hairpin intermediate joins DNA flanking the transposon insertion (Coen et al. 1986). On the basis of our ps1 footprint sequences, we propose a modified “endonuclease model” for Ac excision (see Figure 4). In this model, Ac-mediated cleavage initiates 5′ to the nucleotide immediately flanking the Ac insertion (arrows in Figure 4). Nucleophyllic attack by the free 3′-hydroxyl group joins the top and bottom strands of the flanking DNA, resulting in hairpin formation as recently proposed for Hermes excision (Zhou et al. 2004). The predominant endonucleolytic attack occurs at the nucleotide immediately 3′ to the ligation site and never at the site of ligation (triangles in Figure 4). Repair of the single-stranded gaps followed by ligation of the donor DNA will result in an imperfect 8-bp target-site duplication with transversion mutations at each nucleotide immediately flanking the site of Ac insertion (Figure 4A). Limited exonucleolytic digestion at the single-stranded donor sites followed by ligation of the donor ends will result in 7- or 6-bp insertions (Figure 4, B and C). This model can account for 54 of the 57 ps1 footprint alleles sequenced in this study. The exceptional alleles contain 8- and 9-bp insertions and may have resulted from endonucleolytic cleavage 2 and 3 nucleotides 3′ to the ligation site, respectively. Thus, we propose that endonucleolytic attack of the donor hairpins is never initiated at the site of ligation and occurs rarely at sites more than one nucleotide from the site of ligation. The failure to cleave at the ligation site may reflect a steric hindrance of the transposase molecule or host protein that likely mediates the trans-esterification reaction. A model in which endonucleolytic attack of donor hairpins occurs at sites 3′ to the ligation site is also consistent with the majority of Ds excision products detected at the waxy1 locus by Weil and colleagues (Scott et al. 1996) and with a recent study of Ds transposition in rice (Park et al. 2006). The exceptional cases that are inconsistent with our model may reflect subtle differences in Ac vs. Ds excision or the repair process in somatic vs. gametophytic tissues. Nevertheless, this model predicts that the vast majority of germinal excision alleles recovered from Ac mutagenesis programs will carry imperfect 7- or 8-bp insertions. We are now exploiting this finding to develop “Ac casting” strategies (Singh et al. 2003) that enrich for closely linked Ac transpositions by designing PCR primers that will selectively anneal to these 7- or 8-bp imperfect excision events (L. Conrad, unpublished results).
Figure 4.—
Modified endonuclease model for ps1 footprint formation. Ac-mediated cleavage is indicated by arrows. Curved arrows represent the nucleophylic attack forming DNA hairpins at donor site. Endonuclease cleavage sites are indicated by triangles. Nonhomologous end-joining and repair synthesis results in the formation of an (A) 8-bp insertion with transversions at the junction. Limited exonucleolytic digestion results in (B) 7-bp or (C) 6-bp insertions.
Allelic engineering:
Ps1 encodes lycopene-β-cyclase and is essential for the synthesis of photoprotective carotenoids such as zeaxanthin and for abscisic acid biosynthesis (Singh et al. 2003). Disruption of the Ps1 gene leads to the accumulation of lycopene, photobleaching of seedling tissues, and precocious germination. The maize Ps1 gene is intronless and encodes a predicted 491-amino-acid protein (Singh et al. 2003). An examination of Ac insertion alleles defined domains within the PS1 protein that were critical for activity. Insertions in either orientation within the central coding region of the ps1 gene resulted in a 0% germination rate in our germination assay, presumably due to a failure to splice Ac sequences from the Ps1 transcript or due to transcription termination within Ac, resulting in a truncated PS1 protein. Our finding that all Ac insertions at the 3′-end of Ps1 condition very low germination rates and seedling lethality indicates that the 3′-end of the Ps1 gene is less critical but still indispensable for proper PS1 protein function. The Ac insertions in the 5′-UTR of Ps1 conditioned a very weak mutant phenotype, indicating that a functional PS1 protein was generated in these lines. Thus, despite the absence of introns and the relatively small size of the Ps1 gene (∼2.0 kb), extensive phenotypic variation that was correlated to the position of Ac insertion within the gene was observed. As previously shown at the p1 locus (Athma et al. 1992; Moreno et al. 1992), our findings indicate that Ac will serve as a useful tool in genetic fine-mapping studies to define the boundaries of genes throughout the maize genome.
One powerful application of Ac mutagenesis is to create stable excision alleles for structure–function studies and to introduce novel allelic variation into breeding programs where genetic stability is essential. The predominant Ac excision alleles detected in this study were 7- or 8-bp duplications, resulting in frameshift mutations. This finding is similar to the predominant footprint following Ds excision in somatic tissues of maize (Scott et al. 1996). However, we did recover 6- and 9-bp excision alleles from four independent Ac insertion sites within Ps1. Furthermore, in previous studies of Ds excision, insertions of three, six, and nine nucleotides were recovered at the waxy1, bz1, sh2, and r1 loci (Wessler et al. 1986; Schiefelbein et al. 1988; Giroux et al. 1996; Liu et al. 1996, 1998). As excision events that generate insertions in multiples of three nucleotides will yield in-frame amino acid insertions within the protein-coding region, this class is the most useful in site-directed mutagenesis programs with a goal of creating proteins with slightly altered function. Importantly, all footprint alleles generated through Ac or Ds excision maintain the native context of the altered gene, ensuring similar transcription rates and processing events that are not feasible through the use of transgenics.
In summary, through the use of the genetic scheme outlined in Figure 2, it should be possible to identify germinal Ac excision alleles from any locus where two closely linked Ac insertions are present. As this screen is independent of the phenotype induced by Ac excision, and utilizes a reporter gene that is expressed in the aleurone, a large number of testcross progeny can be easily screened. The ability to screen large numbers of kernels will increase the likelihood of recovery of rare 3-, 6-, and 9-bp duplications and should facilitate site-directed mutagenesis programs utilizing Ac.
Acknowledgments
We thank Liza Conrad, Moira Sheehan, and Kazuhiro Kikuchi for a critical reading of the manuscript and helpful discussions. This work was supported by a grant from the National Science Foundation to T.P.B. (DBI-0076892) and by the Triad Foundation.
References
- Alleman, M., and J. L. Kermicle, 1993. Somatic variegation and germinal mutability reflect the position of transposable element Dissociation within the maize R gene. Genetics 135: 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma, P., E. Grotewold and T. Peterson, 1992. Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics 131: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran, G., C. Echt, T. Bureau and S. Wessler, 1992. Molecular analysis of the maize wx-B3 allele indicates that precise excision of the transposable Ac element is rare. Genetics 130: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R. A., and E. Williams, 1973. Mutable R-Navajo alleles of cyclic origin in maize. Genetics 73: 273–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell, T. P., and L. J. Conrad, 2003. Transposon tagging using Activator (Ac) in maize. Methods Mol. Biol. 236: 157–176. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., and S. L. Dellaporta, 1994. Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell, T. P., B. P. May and S. L. Dellaporta, 1997. The Ac-st2 element of maize exhibits a positive dosage effect and epigenetic regulation. Genetics 147: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, H. G., M. S. Choe, S. H. Lee, S. H. Park, J. C. Koo et al., 1999. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 19: 615–623. [DOI] [PubMed] [Google Scholar]
- Chomet, P., 1988. Characterization of stable and metastable changes of the maize transposable element, Activator. Ph.D. Thesis, State University of New York, Stony Brook, NY.
- Coen, E. S., R. Carpenter and C. Martin, 1986. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell 47: 285–296. [DOI] [PubMed] [Google Scholar]
- Coen, E. S., T. P. Robbins, J. Almeida, A. Hudson and R. Carpenter, 1989. Consequences and mechanism of transposition in Antirrhinum majus, pp. 413–436 in Mobile DNA, edited by D. E. Berg and M. M. Howe. American Society for Microbiology, Washington, DC.
- Conrad, L. J., and T. P. Brutnell, 2005. Ac-immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of Dissociation elements in maize. Genetics 171: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite, M., W. Park, Z. Xu, X. Yan, S. C. Maurais et al., 2002. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, F. X., Jr., B. Pogson, Z. Sun, K. A. McDonald, D. DellaPenna et al., 1996. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8: 1613–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R. K., and M. Freeling, 1990. Clonal analysis of the cell lineages in the male flower of maize. Dev. Biol. 142: 233–245. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S. L., and M. A. Moreno, 1994. Gene tagging with Ac/Ds elements in maize, pp. 219–233 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Dooner, H. K., and A. Belachew, 1989. Transposition pattern of the maize element Ac from the Bz-m2(Ac) allele. Genetics 122: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and J. L. Kermicle, 1971. Structure of the R r tandem duplication in maize. Genetics 67: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisses, J. F., D. Lafoe, L. A. Scott and C. F. Weil, 1997. Novel, developmentally specific control of Ds transposition in maize. Mol. Gen. Genet. 256: 158–168. [DOI] [PubMed] [Google Scholar]
- Emerson, R., 1917. Genetical studies of variegated pericarp in maize. Genetics 2: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux, M. J., J. Shaw, G. Barry, B. G. Cobb, T. Greene et al., 1996. A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 93: 5824–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco, R., P. B. Ouwerkerk, R. J. De Kam, C. Sallaud, C. Favalli et al., 2003. Transpositional behaviour of an Ac/Ds system for reverse genetics in rice. Theor. Appl. Genet. 108: 10–24. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, Modulator, in maize. Genetics 108: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P. D., and V. F. Irish, 2001. The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128: 13–23. [DOI] [PubMed] [Google Scholar]
- Jin, W. Z., S. M. Wang, M. Xu, R. J. Duan and P. Wu, 2004. Characterization of enhancer trap and gene trap harboring Ac/Ds transposon in transgenic rice. J. Zhejiang Univ. Sci. 5: 390–399. [DOI] [PubMed] [Google Scholar]
- Kermicle, J., M. Alleman and S. L. Dellaporta, 1989. Sequential mutagenesis of a maize gene, using the transposable element Dissociation. Genome 31: 712–716. [Google Scholar]
- Kolkman, J., L. J. Conrad, P. R. Farmer, K. Hardeman, K. R. Ahern et al., 2005. Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics 169: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., and C. F. Weil, 2002. The hAT and CACTA superfamily of plant transposons, pp. 565–610 in Mobile DNA, edited by N. L. Craig. American Society for Microbiology Press, Washington, DC.
- Liu, Y. H., M. Alleman and S. R. Wessler, 1996. A Ds insertion alters the nuclear localization of the maize transcriptional activator R. Proc. Natl. Acad. Sci. USA 93: 7816–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. H., L. J. Wang, J. L. Kermicle and S. R. Wessler, 1998. Molecular consequences of Ds insertion into and excision from the helix-loop-helix domain of the maize R gene. Genetics 150: 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, B. P., H. Liu, E. Vollbrecht, L. Senior, P. D. Rabinowicz et al., 2003. Maize-targeted mutagenesis: a knockout resource for maize. Proc. Natl. Acad. Sci. USA 100: 11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1951. Chromosome organization and gene expression. Cold Spring Harbor Symp. Quant. Biol. 16: 13–47. [DOI] [PubMed] [Google Scholar]
- Moreno, M. A., J. Chen, I. Greenblatt and S. L. Dellaporta, 1992. Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics 131: 939–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Neumann, M., J. I. Yoder and P. Starlinger, 1984. The DNA sequence of the transposable element Ac of Zea mays L. Mol. Gen. Genet. 198: 19–24. [Google Scholar]
- Orton, E. R., and R. A. Brink, 1966. Reconstitution of variegated pericarp allele in maize by transposition of Modulator back to P locus. Genetics 53: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov, S., M. Sevugan, Y. De, W. C. Yang, M. Kumaran et al., 1999. Analysis of flanking sequences from Dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. J., H. L. Piao, Y. H. Xuan, S. H. Park, B. I. Je et al., 2006. Analysis of intragenic Ds transpositions and excision events generating novel allelic variation in rice. Mol. Cell 21: 284–293. [PubMed] [Google Scholar]
- Peng, J. R., and N. P. Harberd, 1997. Transposon-associated somatic gai-loss sectors in Arabidopsis. Plant Sci. 130: 181–188. [Google Scholar]
- Pohlman, R. F., N. V. Fedoroff and J. Messing, 1984. The nucleotide sequence of the maize controlling element Activator. Cell 37: 635–643. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J. W., D. B. Furtek, H. K. Dooner and O. E. Nelson, Jr., 1988. Two mutations in a maize bronze-1 allele caused by transposable elements of the Ac-Ds family alter the quantity and quality of the gene product. Genetics 120: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, L., D. LaFoe and C. F. Weil, 1996. Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M., P. E. Lewis, K. Hardeman, L. Bai, J. K. Rose et al., 2003. Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., P. Springer, T. Volpe, S. Haward, J. D. Jones et al., 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Weil, C. F., and R. Kunze, 2000. Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat. Genet. 26: 187–190. [DOI] [PubMed] [Google Scholar]
- Weil, C. F., S. Marillonnet, B. Burr and S. R. Wessler, 1992. Changes in state of the Wx-M5 allele of maize are due to intragenic transposition of Ds. Genetics 130: 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S. R., G. Baran, M. Varagona and S. L. Dellaporta, 1986. Excision of Ds produces waxy proteins with a range of enzymatic activities. EMBO J. 5: 2427–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., X. Li, W. Yuan, G. Chen, A. Kilian et al., 2003. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 35: 418–427. [DOI] [PubMed] [Google Scholar]
- Zhou, L., R. Mitra, P. W. Atkinson, A. B. Hickman, F. Dyda et al., 2004. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 432: 995–1001. [DOI] [PubMed] [Google Scholar]