Abstract
Although the sex-determining gene DMY has been identified on the Y chromosome in the medaka (Oryzias latipes), this gene is absent in most Oryzias species, suggesting that closely related species have different sex-determining genes. Here, we investigated the sex-determination mechanism in O. dancena, which does not possess the DMY gene. Since heteromorphic sex chromosomes have not been reported in this species, a progeny test of sex-reversed individuals produced by hormone treatment was performed. Sex-reversed males yielded all-female progeny, indicating that O. dancena has an XX/XY sex-determination system. To uncover the cryptic sex chromosomes, sex-linked DNA markers were screened using expressed sequence tags (ESTs) established in O. latipes. Linkage analysis of isolated sex-linked ESTs showed a conserved synteny between the sex chromosomes in O. dancena and an autosome in O. latipes. Fluorescence in situ hybridization (FISH) analysis of these markers confirmed that sex chromosomes of these species are not homologous. These findings strongly suggest an independent origin of sex chromosomes in O. dancena and O. latipes. Further analysis of the sex-determining region in O. dancena should provide crucial insights into the evolution of sex-determination mechanisms in vertebrates.
ALMOST all vertebrates have different sexes, males and females. Despite such universal occurrence, sex can be determined by a variety of different mechanisms. Among the major vertebrate groups, highly divergent genetic and environmental factors control sex determination. These include male heterogamety (XX/XY system typified by mammals) and female heterogamety (ZZ/ZW system in birds and snakes), as well as environmental sex determination systems (such as a temperature-dependent sex determination in alligators). In mammals, the sex-determining gene, SRY/Sry has been identified on the Y chromosome (Gubbay et al. 1990; Sinclair et al. 1990). However, no equivalent genes have been found in nonmammalian vertebrates, until recently.
In the medaka, Oryzias latipes, which has an XX/XY sex determination system, DMY was identified as the Y-specific sex-determining gene (Matsuda et al. 2002; Matsuda 2005). This gene encodes a putative transcription factor containing a DM domain, which was originally described as a DNA-binding motif found in two proteins, DSX in Drosophila melanogaster and MAB-3 in Caenorhabditis elegans (Raymond et al. 1998). Vertebrates have several DM domain-containing genes and one of these, DMRT1 (DM-related transcription factor 1) has been implicated in male sexual development in mammals, birds, reptiles, and fishes (Raymond et al. 1999; Smith et al. 1999; De Grandi et al. 2000; Guan et al. 2000; Kettlewell et al. 2000; Marchand et al. 2000). The cDNA sequences of the medaka DMY and DMRT1 showed a high similarity (∼80%) and DMY appears to have arisen through a gene duplication event of an autosomal DMRT1 gene (Matsuda et al. 2002; Nanda et al. 2002).
In contrast to the widespread distribution of Sry in mammals, the DMY gene has not been detected, even in closely related species. Although one sister species of O. latipes has the DMY gene on a homologous Y chromosome (Matsuda et al. 2003), the gene has not been found in other Oryzias species (Kondo et al. 2003). Fishes in the genus Oryzias have been divided into three monophyletic species groups, the latipes, javanicus, and celebensis groups (Takehana et al. 2005) and a recent phylogenetic analysis of DMY and DMRT1 genes from Oryzias species suggested that duplication of the DMRT1 gene (generating the DMY gene) appears to have occurred within the latipes group lineage (Kondo et al. 2004). These findings suggested that Oryzias fishes in other species groups (javanicus and celebensis groups) must have different sex-determining genes. Accordingly, comparisons between closely related medaka fishes with different sex-determination mechanisms should be important in understanding how these diverse developmental mechanisms evolve.
In this study, as a first step to identify a sex-determining gene in other fish species, we investigated the sex-determination system and sex chromosomes in O. dancena, a member of the javanicus group (Takehana et al. 2005). Because a previous cytogenetic study has not reported heteromorphic sex chromosomes in this species (as O. melastigma) (Uwa et al. 1983), a genetic analysis of sex-reversed individuals was performed to demonstrate whether this species has an XX/XY system of sex determination. Moreover, we took advantage of genomic tools established in O. latipes to isolate sex-linked DNA markers, map the sex-determining locus, and identify the sex chromosomes.
MATERIALS AND METHODS
Fish:
All fish used in this study were supplied from a subcenter (Niigata University) of the National BioResource Project (medaka) in Japan. Wild stocks of O. dancena were originally collected at Chidambaram (CB), India, in 1981 and at Phuket (PK), Thailand, in 1988 (for details, see Takehana et al. 2005). Fish were maintained in aquaria under an artificial photoperiod of 14 hr light:10 hr dark at 27° ± 2°.
Sex steroid treatments for sex reversal and progeny testing:
The sex-reversal experiment was performed as previously described by Hamaguchi et al. (2004). Briefly, fertilized eggs of O. dancena (CB) were incubated in water containing estradiol-17β (E2; Sigma, St. Louis) at 0.01, 0.04, and 0.2 μg/ml or methyltestosterone (MT; Sigma) at 0.001, 0.005, and 0.025 μg/ml. Hatched fry were transferred to normal tap water and fed on a commercial pet-food diet until sexual maturation. Sex of the treated fish was judged from secondary sex characteristics and treated fish were subsequently mated with normal fish. Sexing of F1 progeny from the mating was carried out by histological cross-sections of fry gonads, sampled at 20 days after hatching.
Genetic crosses:
By crossing a CB female and (CB female × PK male) an F1 male, 45 backcross progeny were obtained for genotyping. Phenotypic sex was determined by secondary sex characteristics of adult fish and reconfirmed by visual examination of the gonads. We fixed adult fish in 100% ethanol and isolated their genomic DNA from some muscle tissue using the PI-50 isolation system (Kurabo).
Isolation of sex-linked DNA markers and linkage analysis:
We searched for sex-linked markers using expressed sequence tag (EST) markers mapped to each of the O. latipes chromosomes [linkage group (LG)] (Naruse et al. 2004). ESTs were amplified using previously published primers designed for O. latipes (supplemental Table 1 at http://www.genetics.org/supplemental/). PCR genotyping for two loci, BJ014360 and BJ732639, was performed using primers that we designed on the basis of the EST sequences (http://mbase.bioweb.ne.jp/∼dclust/medaka_top.html) and the draft genome sequence (http://dolphin.lab.nig.ac.jp/medaka/) of O. latipes. PCR amplification of each EST marker was performed in a total volume of 10 μl for 3 min at 95° followed by 35 cycles of 10 sec at 95°, 30 sec at 55°–60°, 60 sec at 72°, with a final elongation step of 3 min at 72°. Restriction fragment length polymorphisms (RFLPs) in PCR-amplified fragments of the parents (CB female and PK male) were analyzed by polyacrylamide gel electrophoresis. For those markers showing a polymorphic pattern between the parents, F1 and backcross progeny were genotyped and assessed as to whether such polymorphisms segregated with phenotypic sex. Using these isolated sex-linked ESTs, a sex-linkage map of O. dancena was constructed.
Fluorescence in situ hybridization analysis with fosmid and BAC clones:
A fosmid genome library of O. dancena was constructed from an F1 male between CB and PK using the Copy Control Fosmid Library Production kit (Epicentre, Madison, WI). Fosmid clones containing sex-linked ESTs were screened by colony hybridization. A fosmid clone Od38_01 (containing BJ014360) was used as a probe. A bacterial artificial chromosome (BAC) genomic library, constructed from the Hd-rR strain of O. latipes (Matsuda et al. 2001), was also screened and three clones, Md0173J11 (containing SL1), Md0172B19 (containing DMRT1), and Md0172I07 (containing OLd17.11a), were isolated. These BAC clones were located, respectively, on the sex chromosomes (LG 1) and on autosomes (LG 9 and LG 10) in O. latipes.
Metaphase cells from cultured caudal fins were prepared by standard cytogenetic methods (Uwa and Ojima 1981; Matsuda et al. 1998). Fluorescence in situ hybridization (FISH) was performed as described (Matsuda and Chapman 1995). Probe DNAs of the genomic clones were labeled separately by nick translation using biotin-16dUTP (Roche, Indianapolis) or digoxigenin-11-dUTP (Roche). For two-color hybridization, equal amounts of labeled probes and a 500-fold amount of autoclaved genomic DNA of O. latipes were mixed with hybridization solution and preannealed for 30 min at 37°. After overnight hybridization, probes were detected with avidin–FITC (Roche) and rhodamine-labeled anti-digoxigenin antibodies (Roche). Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and examined under a Nikon Eclipse 80i microscope using three filters (UV-1A, B-2A, and G-2A). Images were captured with a DXM1200C digital camera (Nikon, Tokyo).
RESULTS
Progeny testing of sex-reversed fish:
Sex ratios of the hormone-treated fish are shown in Table 1. The sex ratio of the MT-treated group was biased toward male, suggesting that MT induced sex reversal from female to male. In the groups reared under normal conditions, the sex ratio was almost 1:1 (data not shown). Therefore, the MT-treated group was expected to include sex-reversed males whose genetic sex is female (XX or ZW). On the other hand, the sex ratio of the E2-treated groups did not deviate toward females, suggesting that E2 could not reverse the sex of the fish from male to female at concentrations of 0.01–0.04 μg/ml. In addition, all fish died at a concentration of 0.2 μg/ml E2 before or just after hatching (data not shown).
TABLE 1.
Sex ratio of the hormone-treated fish
| Concentration (μg/ml) | Male | Female | Total |
|---|---|---|---|
| Methyltestosterone | |||
| 0.001 | 20 | 2 | 22 |
| 0.005 | 30 | 5 | 35 |
| 0.025 | 20 | 0 | 20 |
| Estradiol-17β | |||
| 0.01 | 10 | 12 | 22 |
| 0.04 | 10 | 9 | 19 |
Of 20 mature males obtained by the 0.025 μg/ml MT treatment, 11 were randomly selected and subjected to a progeny test by matings with normal females. The sex ratio of the offspring from each mating is shown in Table 2. As a result, 5 of 11 males from the MT-treated group yielded all-female progeny, demonstrating that these males were sex-reversed XX males. This result reveals that O. dancena has a male-heterogametic (XX/XY) sex-determination system.
TABLE 2.
Sex ratio of the offspring of mating between a methyltestosterone-treated male and a normal female
| Mating no. | Male | Female | Total | Remark |
|---|---|---|---|---|
| 1 | 0 | 8 | 8 | Sex-reversed XX male |
| 2 | 18 | 18 | 36 | |
| 3 | 17 | 16 | 33 | |
| 4 | 9 | 14 | 23 | |
| 5 | 16 | 21 | 37 | |
| 6 | 15 | 13 | 28 | |
| 7 | 0 | 43 | 43 | Sex-reversed XX male |
| 8 | 14 | 19 | 33 | |
| 9 | 0 | 47 | 47 | Sex-reversed XX male |
| 10 | 0 | 47 | 47 | Sex-reversed XX male |
| 11 | 0 | 57 | 57 | Sex-reversed XX male |
Sex-linkage map of O. dancena:
To isolate sex-linked DNA markers in O. dancena, we searched for RFLPs between the parents using ESTs established in O. latipes and genotyped F1 and backcross progeny. We screened 397 ESTs and identified 65 polymorphic markers. Linkage analysis showed that 8 of these 65 markers segregated with the phenotypic sex. The female parent was homozygous, and the male parent and/or F1 male were heterozygous in these markers. In the backcross progeny, males had the paternal genotype, while females had the maternal genotype, confirming an XX/XY sex determination system (Figure 1).
Figure 1.—
Linkage analysis of EST markers in Oryzias dancena. Electrophoretic patterns of MF01SSA032H09 PCR product digested with HinfI (A) and BJ014360 PCR product digested with HhaI (B) are shown. Genomic DNA from female (F) and male (M) fish was used as templates for PCR. Abbreviations: P, parents (CB female and PK male); F1, F1 progeny from the parents; BC1, backcross progeny. Note that the digested bands (arrowheads) segregated perfectly with BC1 males.
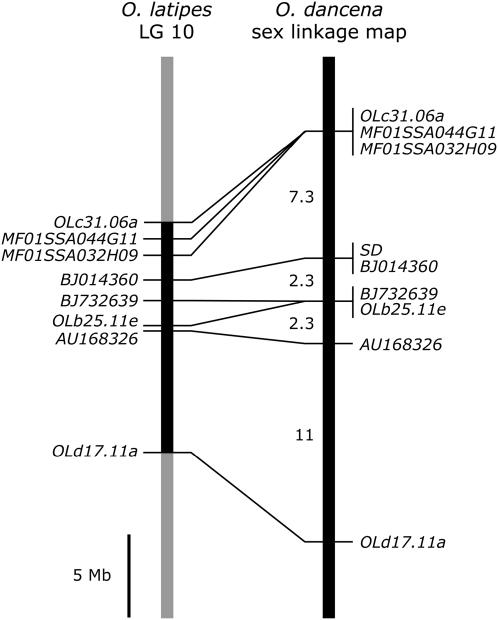
Using eight isolated EST markers, we constructed a sex-linkage map of O. dancena and showed that the sex-determining (SD) locus was mapped to the same position with an EST, BJ014360 (Figure 2). On the basis of the draft genome sequence data of O. latipes (http://dolphin.lab.nig.ac.jp/medaka/), all eight ESTs were located on LG 10 in O. latipes. Comparison of gene order between the O. dancena sex-linkage map and the physical map of O. latipes LG 10 indicated a conserved synteny between the two chromosomes. These results suggest that the sex chromosomes in O. dancena are homologous to an autosome (LG 10) of O. latipes, whose sex chromosomes are LG 1.
Figure 2.—
Comparison of gene order between a sex-linkage map in Oryzias dancena and a physical map of LG 10 in O. latipes. Lines between the compared chromosomes connect positions of orthologous gene pairs in these two species. The distances between franking markers are shown in physical length (left) and in centimorgans (right). Map positions for genes and distances in O. dancena were derived from this study; those in O. latipes were obtained from the draft genome sequence data (http://dolphin.lab.nig.ac.jp/medaka/).
In this sex-linkage map, the total map length was ∼23 cM in male meiosis. The corresponding region of O. latipes has a similar length (24.4 cM) (http://dolphin.lab.nig.ac.jp/medaka/), suggesting that recombination along the sex chromosomes is not suppressed in O. dancena. Indeed, we obtained 4/45 (8.9%) recombinants between MF01SSA032H09 and BJ732639, which flanked the SD locus. On the basis of the draft genome sequence, this region was calculated to be ∼3.0 Mb in O. latipes. These findings suggest a high recombination frequency around the sex-determining region in O. dancena.
Identification of sex chromosomes by FISH mapping:
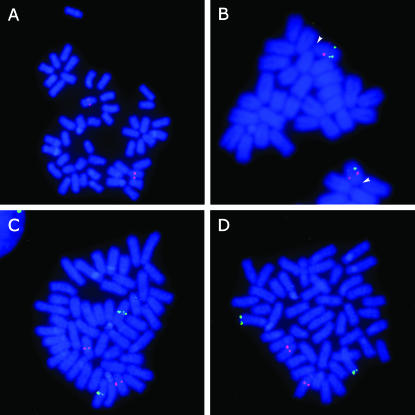
The karyotype of O. dancena consists of 24 acrocentric chromosome pairs with no heteromorphic sex chromosomes (Uwa et al. 1983). To identify the sex chromosomes in O. dancena, we conducted a FISH analysis of male chromosomes using a fosmid clone containing BJ014360 as a probe, which is tightly linked to the SD locus. Clear hybridization signals were observed on the middle position of an acrocentric chromosome pair, identifying the cryptic sex chromosomes (Figure 3A). We also found sex chromosome localization of the O. latipes BAC clone (Md0172I07) containing another sex-linked EST, OLd17.11a. Two-color FISH indicated that the BJ014360 signal was located more proximal to the centromere than the OLd17.11a signal on the sex chromosomes (Figure 3B). Morphological differences between the X and Y chromosomes were not observed in this study, indicating that O. dancena has homomorphic sex chromosomes.
Figure 3.—
FISH analysis of male metaphase chromosomes in Oryzias dancena using O. dancena fosmid and O. latipes BAC clones. (A) FISH mapping of a fosmid clone (Od38_01) containing a sex-linked EST, BJ014360. A specific hybridization signal (red) was located on a homomorphic acrocentric chromosome pair. (B) Gene ordering of the sex-linked ESTs, BJ014360 (fosmid Od38_01) and OLd17.11a (BAC Md0172I07), on the sex chromosomes. The locations of the markers are visualized as red and green signals, respectively. Arrowheads indicate the centromere positions. (C) Chromosomal location of the O. latipes sex-chromosomal marker SL1 (BAC Md0173J11, green) and the O. dancena sex-chromosomal marker BJ014360 (fosmid Od38_01, red). (D) Chromosomal location of the DMRT1 gene (BAC Md0172B19, green) with the BJ014360 fosmid, Od38_01 (red).
On the other hand, the BAC clone (Md0173J11), containing an O. latipes sex-chromosomal marker (SL1), was not located on the sex chromosomes in O. dancena (Figure 3C), confirming that sex chromosomes are different between these two species. Furthermore, the hybridization signals of the DMRT1-positive BAC (Md0172B19) were detected only on a pair of autosomes (Figure 3D). In O. latipes, previous FISH analyses revealed the presence of DMRT1 at the tip of a pair of autosomes, as well as at one of the sex chromosome pair (namely the DMY gene on the Y chromosomes) (Kondo et al. 2004, 2006). Although we found a similar FISH pattern to that of autosomal DMRT1 in O. latipes, the additional Y-chromosomal location (corresponding to the DMY) was not observed in O. dancena. Hence, these results strongly suggest that Y chromosomes of O. dancena and O. latipes are not homologous.
DISCUSSION
O. dancena has an XX/XY sex-determination system:
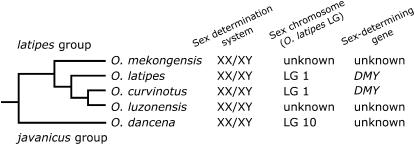
It has been demonstrated that, on the basis of sex ratios in the progeny of sex-reversed fish and segregation patterns of sex-linked DNA markers, O. dancena in the javanicus species group has a male-heterogametic (XX/XY) sex-determination system. A previous study showed that all four fishes in the latipes group also have an XX/XY sex-determination system (Hamaguchi et al. 2004) (Figure 4). Two of these four species, O. latipes and O. curvinotus, have the common sex-determining gene, DMY, on the homologous Y chromosomes (Matsuda et al. 2002, 2003). On the other hand, the gene has not been detected in the other two species, O. luzonensis and O. mekongensis (Kondo et al. 2003, 2004). On the basis of these findings with a phylogenetic analysis of DMY and DMRT1, Kondo et al. (2004) suggested that the DMY gene appears to have occurred in the common ancestor of O. latipes, O. curvinotus, and O. luzonensis. Therefore, the DMY gene is considered to be recently lost in O. luzonensis and originally absent in O. mekongensis and in other Oryzias species, such as O. dancena. This suggests that different sex-determining genes have evolved in Oryzias species, although male heterogamety is likely to be common in the genus.
Figure 4.—
Phylogenetic relationships and sex-determination mechanisms in Oryzias fishes. The phylogenetic information was taken from Takehana et al. (2005). Data of sex-determination mechanisms in O. latipes, O. curvinotus, O. luzonensis, and O. mekongensis were obtained from Matsuda et al. (2002, 2003) and Hamaguchi et al. (2004).
Identification of sex chromosomes in O. dancena:
Oryzias species with no DMY gene must have different sex chromosomes from those in O. latipes; however, no sex chromosomes have been identified in these species so far. In this study, we screened sex-linked markers of O. dancena using ESTs established in O. latipes. As a result, most ESTs primers worked well in O. dancena, and we could successfully isolate eight sex-linked EST markers. Linkage analysis showed that these ESTs are located on an autosome (LG 10) in O. latipes with the same gene order, indicating a conserved synteny between sex chromosomes in O. dancena and an autosome of O. latipes. In addition, FISH analysis confirmed that sex chromosomes in O. dancena and O. latipes were not homologous. These results indicated an independent origin of X/Y chromosomes in these two species, suggesting that a novel sex-determining gene is located on the sex chromosomes in O. dancena.
The lack of conservation of sex chromosomes among closely related fish species is probably common. Although all salmonid fishes have a male heterogametic system, closely related species have evolved different sex chromosomes, as evidenced by a comparative linkage analysis (Woram et al. 2003) and FISH analyses (Phillips et al. 2001, 2005). In sticklebacks, Gasterosteus aculeatus and G. wheatlandi also appear to have different X/Y sex chromosomes, and Apeltes quadracus has heteromorphic Z/W sex chromosomes (Peichel et al. 2004). These studies suggest that fishes may use a variety of sex-determining genes.
Sex-determining region in O. dancena:
Linkage analysis mapped the SD locus between MF01SSA032H09 and BJ732639 and showed that BJ014360 is tightly linked to the SD locus with no recombinants (0/45). This suggests that a single chromosomal region controls sex in O. dancena. FISH analysis indicated that the sex-determining region is located on the middle of one acrocentric chromosome pair, in accordance with the linkage analysis. Therefore, only this region should be different between X and Y chromosomes.
Standard models of sex chromosome evolution hypothesized that the first step is the occurrence of a novel single locus on an autosome, in which heterozygosity leads to the development of one sex and homozygosity to the other sex, thereby establishing a protosex chromosome system. Heteromorphic sex chromosomes are considered to arise through the recombination isolation between such homologous sex chromosomes. This suppression of recombination maintained one chromosome (such as Y) in a constant heterozygous state in one sex and the subsequent degeneration process spreads the sex-specific region over almost the entire chromosome, as in mammals (reviewed in Graves 2006).
Our results indicate that sex chromosomes in O. dancena are at an early stage of evolution, as in O. latipes. First, phenotypic sex was easily converted by sex hormones, and the resultant sex-reversed fish were fully fertile. Furthermore, YY males have been obtained from estrogen-induced XY females (Y. Takehana and D. Demiyah, unpublished results), indicating viability of YY individuals. Second, sex-chromosomal crossing over occurred over almost the entire length of the chromosome, indicating that this recombining section is considered to be a pseudoautosomal region. In addition, the region between MF01SSA032H09 and BJ732639, which flanked the SD locus in O. dancena, was calculated to be 3.0 Mb in O. latipes, on the basis of the draft genome sequence. This suggests that the Y-specific region of O. dancena is likely to be very small. Finally, FISH analysis demonstrated that the Y chromosome is not cytogenetically distinguished from the X, although the sex-determination system in the species is male heterogametic. Taken together, these findings suggest that there are no functional differences between X and Y sex chromosomes, other than the sex-determining gene.
Only two sex-determining genes, Sry and DMY, have been identified in vertebrates so far. In this study, we have shown that O. dancena should have a novel sex-determining gene on the sex chromosomes homologous to an autosome (LG 10) in O. latipes. One approach to identify sex-determining genes is positional cloning. This method has been successfully used to isolate human and medaka sex-determining genes. Two major conditions are required for using this method: a firm genetic sex determination and a feasibility of genetic mapping for the SD locus (Matsuda 2005). O. dancena has a strict genetic sex determination (XX/XY system) and a high recombination frequency around the SD locus, satisfying both requirements. In addition, O. dancena and O. latipes share a similar character of sex chromosomes, a very small Y-specific region, suggesting that this method will also be effective in O. dancena. Construction of a high-resolution recombination map around the SD locus and chromosome walking to the SD locus are necessary steps to isolate the sex-determining gene using the positional cloning method. This is currently in progress. Comparative analyses of sex-determining genes and sex chromosomes among Oryzias species will prove useful in unraveling the evolution of sex-determination mechanisms in vertebrates.
Acknowledgments
We thank Yoichi Matsuda, Takahiro Murakami (Hokkaido University), and Masaru Matsuda (National Institute for Basic Biology) for their helpful technical advice on FISH analysis and Wichian Magtoon (Srinakharinwirot University) for his generous help in the collection of materials. This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (16370094) to M.S.
References
- De Grandi, A., V. Calvari, V. Bertini, A. Bulfone, G. Peverali et al., 2000. The expression pattern of a mouse doublesex-related gene is consistent with a role in gonadal differentiation. Mech. Dev. 90: 323–326. [DOI] [PubMed] [Google Scholar]
- Guan, G., T. Kobayashi and Y. Nagahama, 2000. Sexually dimorphic expression of two types of DM (Doublesex/Mab-3)-domain genes in a teleost fish, the Tilapia (Oreochromis niloticus). Biochem. Biophys. Res. Commun. 272: 662–666. [DOI] [PubMed] [Google Scholar]
- Gubbay, J., J. Collignon, P. Koopman, B. Capel, A. Economou et al., 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346: 245–250. [DOI] [PubMed] [Google Scholar]
- Graves, J. A. M., 2006. Sex chromosome specialization and degeneration in mammals. Cell 124: 901–914. [DOI] [PubMed] [Google Scholar]
- Hamaguchi, S., Y. Toyazaki, A. Shinomiya and M. Sakaizumi, 2004. The XX–XY sex-determination system in Oryzias luzonensis and O. mekongensis revealed by the sex ratio of the progeny of sex-reversed fish. Zool. Sci. 21: 1015–1018. [DOI] [PubMed] [Google Scholar]
- Kettlewell, J. R., C. S. Raymond and D. Zarkower, 2000. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis 26: 174–178. [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, S. Asakawa, N. Shimizu et al., 2003. Absence of the candidate male sex-determining gene dmrt1b(Y) of medaka from other fish species. Curr. Biol. 13: 416–420. [DOI] [PubMed] [Google Scholar]
- Kondo, M., I. Nanda, U. Hornung, M. Schmid and M. Schartl, 2004. Evolutionary origin of the medaka Y chromosome. Curr. Biol. 14: 1664–1669. [DOI] [PubMed] [Google Scholar]
- Kondo, M., U. Hornung, I. Nanda, S. Imai, T. Sasaki et al., 2006. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 16: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand, O., M. Govoroun, H. D'Cotta, O. McMeel, J. Lareyre et al., 2000. DMRT1 expression during gonadal differentiation and spermatogenesis in the rainbow trout, Oncorhynchus mykiss. Biochim. Biophys. Acta 1493: 180–187. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., 2005. Sex determination in the teleost medaka, Oryzias latipes. Annu. Rev. Genet. 39: 293–307. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., C. Matsuda, S. Hamaguchi and M. Sakaizumi, 1998. Identification of the sex chromosomes of the medaka, Oryzias latipes, by fluorescence in situ hybridization. Cytogenet. Cell Genet. 82: 257–262. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., N. Kawato, S. Asakawa, N. Shimizu, Y. Nagahama et al., 2001. Construction of a BAC library derived from the inbred Hd-rR strain of the teleost fish, Oryzias latipes. Genes Genet. Syst. 76: 61–63. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., T. Sato, Y. Toyazaki, Y. Nagahama, S. Hamaguchi et al., 2003. Oryzias curvinotus has DMY, a gene that is required for male development in the medaka, O. latipes. Zool. Sci. 20: 159–161. [DOI] [PubMed] [Google Scholar]
- Matsuda, Y., and V. M. Chapman, 1995. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16: 261–272. [DOI] [PubMed] [Google Scholar]
- Nanda, I., M. Kondo, U. Hornung, S. Asakawa, C. Winkler et al., 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse, K., M. Tanaka, K. Mita, A. Shima, J. Postlethwait et al., 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel, C. L., J. A. Ross, C. K. Matson, M. Dickson, J. Grimwood et al., 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Phillips, R. B., N. R. Konkol, K. M. Reed and J. D. Stein, 2001. Chromosome painting supports lack of homology among sex chromosomes in Oncorhynchus, Salmo, and Salvelinus (Salmonidae). Genetica 111: 119–123. [DOI] [PubMed] [Google Scholar]
- Phillips, R. B., M. R. Morasch, L. K. Park, K. A. Naish and R. H. Devlin, 2005. Identification of the sex chromosome pair in coho salmon (Oncorhynchus kisutch): lack of conservation of the sex linkage group with chinook salmon (Oncorhynchus tshawytscha). Cytogenet. Genome Res. 111: 166–170. [DOI] [PubMed] [Google Scholar]
- Raymond, C. S., C. E. Shamu, M. M. Shen, K. J. Seifert, B. Hirsch et al., 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391: 691–695. [DOI] [PubMed] [Google Scholar]
- Raymond, C. S., J. R. Kettlewell, B. Hirsch, V. J. Bardwell and D. Zarkower, 1999. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev. Biol. 215: 208–220. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths et al., 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240–244. [DOI] [PubMed] [Google Scholar]
- Smith, C. A., P. J. McClive, P. S. Western, K. J. Reed and A. H. Sinclair, 1999. Conservation of a sex-determining gene. Nature 402: 601–602. [DOI] [PubMed] [Google Scholar]
- Takehana, Y., K. Naruse and M. Sakaizumi, 2005. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 36: 417–428. [DOI] [PubMed] [Google Scholar]
- Uwa, H., and Y. Ojima, 1981. Detailed and banding karyotype analysis of the medaka, Oryzias latipes in cultured cells. Proc. Jpn. Acad. 57B: 39–43. [Google Scholar]
- Uwa, H., T. Iwamatsu and O. P. Saxena, 1983. Karyotype and cellular DNA content of the Indian ricefish, Oryzias melastigma. Proc. Jpn. Acad. 59B: 43–47. [Google Scholar]
- Woram, R. A., K. Gharbi, T. Sakamoto, B. Hoyheim, L. E. Holm et al., 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]