Abstract
The yeast spindle pole body (SPB) plays a unique role in meiosis, initiating both spindle assembly and prospore membrane synthesis. SPO1, induced early in development, encodes a meiosis-specific phospholipase B (PLB) homolog required at three stages of SPB morphogenesis: MI, MII, and spore formation. Here we report in-depth analysis of the SPO1 gene including its transcriptional control by regulators of early gene expression, protein localization to the ER lumen and periplasmic space, and molecular genetic studies of its role in meiosis. Evidence is presented that multiple arrest points in spo1Δ occur independently, demonstrating that Spo1 acts at distinct steps. Loss of Spo1 is suppressed by high-copy glycosylphosphatidylinositol (GPI) proteins, dependent on sequence, timing, and strength of induction in meiosis. Since phosphatidylinositol (PI) serves as both an anchor component and a lipase substrate, we hypothesized that GPI-protein expression might substitute for Spo1 by decreasing levels of its potential substrates, PI and phosphatidylinositol phosphates (PIPs). Partial spo1Δ complementation by PLB3 (encoding a unique PLB capable of cleaving PI) and relatively strong Spo1 binding to PI(4)P derivatives (via a novel N-terminal lysine-rich fragment essential for Spo1 function) are consistent with this view. Epistasis of SPO1 mutations to those in SPO14 (encoding a PLD involved in signaling) and physical interaction of Spo1 with Spo23, a protein regulating PI synthesis required for wild-type sporulation, further support this notion. Taken together these findings implicate PI and/or PIPs in Spo1 function and suggest the existence of a novel Spo1-dependent meiosis-specific signaling pathway required for progression of MI, MII, and spore formation via regulation of the SPB.
THE unicellular eukaryote Saccharomyces cerevisiae undergoes gametogenesis in response to both cell type and environmental signals (for recent reviews see Nasmyth 2003; Esposito 2006). A number of genes are specifically required for this process that are not expressed during vegetative growth (Chu et al. 1998; Primig et al. 2000). Premeiotic DNA synthesis, followed by genetic recombination and two successive nuclear divisions (MI and MII), results in the formation of asci containing four haploid meiotic products encapsulated in spores. The process of spore wall development is highly coordinated with progression of the meiotic divisions beginning at approximately the same time as MII. The yeast spindle pole body (SPB) plays a unique role in meiosis, initiating both spindle assembly and prospore membrane synthesis. The SPB is a tripartite structure embedded in the nuclear membrane (reviewed in Jaspersen and Winey 2004). During early MII, SPBs undergo a modification enlarging their outer plaques, making them distinct from both mitotic and MI SPBs (Moens 1974). Several meiosis-specific proteins localize to the modified outer plaques (MOPs) and promote recruitment of lipid vesicles that fuse to form an expanding prospore membrane (Knop and Strasser 2000; Moreno-Borchart et al. 2001; Neiman 2005). This process is controlled by a specialized branch of the secretory pathway (Neiman 1998). Prospores then mature by the ordered assembly of mannan, glucan, chitosan, and dityrosine layers, respectively (Smits et al. 2001; Coluccio et al. 2004). Since SPBs nucleate both spindle formation (at MI and MII) and prospore membrane synthesis, regulation of its morphogenesis is a likely target of processes ensuring coordination of the nuclear divisions with gamete differentiation. At present, little is known about the mechanism(s) coordinating meiosis with gamete development in any organism. This study aims at understanding this process, using budding yeast as a model system.
We previously provided evidence that SPO1, a gene implicated in meiotic SPB morphogenesis, potentially functions in coordinating the divisions with gamete development (Tevzadze et al. 2000). SPO1 is required at three stages of meiosis where proper SPB morphogenesis is essential: SPB duplication at MI and MII and at spore formation. It is predicted to encode a phospholipase B (PLB) homolog (Tevzadze et al. 1996) induced early in meiosis and expressed through ascus formation, consistent with its requirement for proper SPB function at successive steps of development. A conserved serine within the lipase active site [essential for PLB biochemical activity (Sharp et al. 1994)] is required for its meiotic function (Tevzadze et al. 2000). Unlike the other known budding yeast PLB enzymes, Plb1, Plb2, Plb3 (Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999), and Nte1 (Zaccheo et al. 2004), Spo1 is the only one that is meiosis specific.
In initial genetic studies we found that the spo1 sporulation defect is partially suppressed by high-copy CWP1 (Tevzadze et al. 2000). This gene encodes a cell wall protein with a C-terminal segment characteristic of glycosylphosphatidylinositol (GPI)-anchored proteins (Shimoi et al. 1995). GPIs are inositol-containing glycolipids, which covalently attach these proteins to the cell wall or external surface of the plasma membrane in mammals, yeast, and protozoa (Ikezawa 2002; Mayor and Riezman 2004). Some phospholipases, including Plb1 and Plb2 (Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999), but not Spo1, are anchored this way. The structure of the GPI anchor is well conserved in all eukaryotes. Here we report spo1 suppression by two other GPI proteins, Spo19 and Cwp2, dependent on level and timing of expression, and provide new evidence supporting the idea that the putative Spo1 lipase likely acts on phosphatidylinositol (PI) (or its phosphorylated derivatives) in a novel meiosis-specific signaling pathway coordinating successive stages of meiosis.
MATERIALS AND METHODS
Strains and plasmids:
Escherichia coli strains DH5α and KC8 were employed for propagation and maintenance of plasmid DNA and BL21(DE3) for bacterial expression of yeast proteins. The S. cerevisiae strains used in this study are listed in Table 1.
TABLE 1.
Yeast strains
| Strain | Relevant genotype | Source |
|---|---|---|
| REE2221 | MATahis2 leu1-12 spo1-1 ura3-1 | S. Klapholz (this lab) |
| REE945 | MATα ade2 can1 cyh2 his7 leu1-c lys2 met13-c spo1-1 trp5 tyr1 ura3-1 | S. Klapholz (this lab) |
| W303-1A; W303-1B | MATaade2 can1-100r his3-11,15 leu2-3,112 trp1-1 ura3-1; isogenic MATα | R. Rothstein (Columbia University) |
| GTY361; GTY362 | W303-1A; W303-1B spo1Δ∷KanMX6 | This work |
| GTY297; GTY298 | W303-1A; W303-1B spo14Δ∷KanMX6 | This work |
| GTY118; GTY119 | W303-1A; W303-1B spo19Δ∷KanMX6 | This work |
| GTY196; GTY197 | W303-1A; W303-1B spo23Δ∷KanM6 | This work |
| GTY274; GTY275 | W303-1A; W303-1B plb3Δ∷KanMX6 | This work |
| GTY286; GTY287 | W303-1A; W303-1B cwp1Δ∷KanMX6 | This work |
| GTY90; GTY91 | W303-1A; W303-1B spo1Δ∷HIS3 | Tevzadze et al. (2000) |
| GTY157; GTY158 | GTY90; GTY91 URA3:SPO1-6xMYC:ura3-1 | This work |
| GTY114; GTY115 | GTY90; GTY91 cwp1Δ∷URA3 | This work |
| GTY247; GTY248 | GTY90; GTY91 plb3Δ∷KanMX6 | This work |
| GTY321; GTY322 | GTY157; GTY158 SPO23-FLAG:KanMX6 | This work |
| GTY349; GTY350 | GTY90; GTY91 TRP1:HDEL-GFP:trp1-1 LEU2:TPI1p-SPO1-6xMYC:leu2-3,112 | This work |
| GTY357; GTY358 | GTY90; GTY91 TRP1:HDEL-GFP:trp1-1 LEU2:SPO1p-SPO1-6xMYC:leu2-3,112 | This work |
| yC66 | MATacan1-100 his3-11,15 leu1-12 lys2-1 trp1-1 tyr1-1 ura3-1 | Steber and Esposito (1995) |
| yC67 | MATα cyh2 his3-11,15 leu1-c met13-c trp1-1 tyr1-2 ura3-1 | Steber and Esposito (1995) |
| YAH67; YAH68 | yC66; yC67 ume4Δ∷HIS3 | Ann Helms (this lab) |
| yC105; yC106 | yC66; yC67 ume6Δ | Steber and Esposito (1995) |
| GTY52; GTY53 | yC66 ; yC67 spo14Δ∷URA3 | This work |
| GTY34; GTY35 | yC66; yC67 ime1Δ∷URA3 | This work |
| GTY54; GTY55 | GTY52; GTY53 spo1Δ∷TRP1 | This work |
| GTY61; GTY62 | yC66; C67 cwp1Δ∷URA3 | This work |
| GTY68; GTY69 | yC66; yC67 spo1Δ∷HIS3 | This work |
| GTY71; GTY72 | GTY68; GTY69 URA3:urs1-SPO1:ura3-1 | This work |
| GTY73; GTY74 | GTY68; GTY69 URA3:uash-SPO1-ura3-1 | This work |
| GTY112; GTY113 | GTY68; GTY69 cwp1Δ∷URA3 | This work |
| GTY203; GTY204 | yC66; yC67 spo1Δ∷KanMX6 | This work |
| GTY218; GTY219 | yC66; yC67 spo19Δ∷KanMX6 | This work |
| NSY72 | MATahis3-Δ200 lys2-801 sec14-1 ura3-52 | Nava Segev (University of Illinois, Chicago) |
| NSY73 | MATα ade2-1Δ 1 his3-Δ200 trp1-Δ sec14-1 ura3-52 | Nava Segev (University of Illinois, Chicago) |
| GTY200; GTY201 | NSY72; NSY73 spo1Δ∷KanMX6 | This work |
| B55 | MATaade2-101 his4-519 leu2-3,112 sec14-1 ts | V. Bankaitis (University of North Carolina) |
| GTY466 | B55 LEU2:YIplac-128T:leu2-3,112 | This work |
| GTY467 | B55 LEU2:TPI1p-SPO1:leu2-3,112 | This work |
Plasmids for complementation:
pGT106 contains SPO1 (on a 3.5-kb BamHI fragment) and pGT42 contains CWP1 (on a 1.8-kb XhoI + EcoRI fragment), cloned into a URA3 high-copy plasmid pRS426 (Christianson et al. 1992). pGT106 fully complements both spo1-1 ts and spo1Δ, forming >60% spores in the W303 background (Tevzadze et al. 2000). pGT118 and pGT119 contain the SPO1 ORF tagged with six copies of the myc epitope at the C terminus immediately before the STOP codon cloned into pRS426 (Christianson et al. 1992) and pRS306, a URA3-marked integrative plasmid (Sikorski and Hieter 1989), respectively. pGT119 was linearized by StuI to direct integration into the ura3-1 locus. The tagged construct complements spo1 at levels comparable to the untagged locus (>50% asci). pS25, carrying a 1.0-kb SalI internal fragment of SPO14 cloned into pRS306, was used to create a SPO14 gene disruption/deletion marked with URA3 (Honigberg et al. 1992). pGT34, used to construct a TRP1-marked deletion of SPO1, and pGT43, used to make a URA3-marked deletion of CWP1, were described previously (Tevzadze et al. 2000).
ER targeting and Spo1 localization plasmids:
YIplac128-T and YIp-lac204/TKC-GFP-HDEL were generously provided by B. S. Glick (University of Chicago). YIplac128-T, an integrative plasmid carrying LEU2, contains a strong constitutive promoter from TPI1, which encodes triose-phosphate isomerase. pJP19 and pJP22 contain an untagged SPO1 ORF from pGT106 and the SPO1-6MYC ORF from pGT118, respectively, cloned into the SacI–BamHI sites of YIplac128-T under the control of the TPI1 promoter (the SacI sites were engineered immediately upstream of the SPO1 ATG). These plasmids were linearized by ClaI for integration into leu2-3,114. The YIp-lac204/TKC-GFP-HDEL plasmid, marked by TRP1, contains a GFP insert. The GFP coding sequence was modified by adding the N-terminal 135-bp sequence from the KAR2 ORF (for targeting to the ER lumen) and the C-terminal sequence encoding HDEL, a tetrapeptide that ensures retention of GFP in the ER lumen (Rossanese et al. 2001). This plasmid was linearized with EcoRV for integration into TRP1.
Mutated SPO1 promoter plasmids:
The “urs-SPO1” allele contains multiple mutations disrupting two potential overlapping URS1 core sequences (GGCGGC), one of which starts at +268 and the other at +271 (see Figure 2A, results). The mutations were introduced at positions +268, +269, +271, +272, +274, +275, and +276 (G–A transitions) and +270 and +273 (C–T transitions). This allele was cloned into the high-copy plasmid pRS426 and into the integrating vector pRS306 to create pGT62 and pGT64, respectively. The “uasH-SPO1” allele also contains multiple mutations in the UASH sequence (227GCGTGTTGAAAG238) at positions +227 (G–C), +229 (G–T), +236 (A–T), +237 (A–C), and +238 (G–T). This allele was also cloned into pRS426 and pRS306 to construct plasmids pGT63 and pGT65, respectively. Mutations in both URS1 and UASH cause a loss of spo1 complementation.
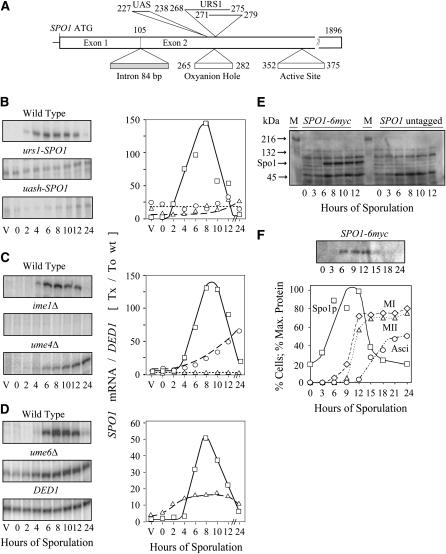
Figure 2.—
Analysis of SPO1 transcription during meiosis. (A) Localization of the URS1 and UASH sites within the SPO1 ORF. (B–D) S1 nuclease protection assays of SPO1 expression. SPO1 transcript levels were normalized to the DED1 transcript at each time point (Tx) and to the initial (To) value upon transfer to sporulation medium. Transcription in isogenic diploids is shown for (B) wild-type (yC66/yC67, squares), mutated URS1 (circles), and UASH (triangles) sequences in the SPO1 promoter; (C) wild-type (yC66/yC67, squares), ime1 (triangles), and ume4/sin3 (circles); and (D) wild type (SFY59/yC67, squares) and ume6 (triangles). (E) Western analysis of total protein samples during sporulation at 0, 3, 6, 8, 10, and 12 hr: left, a/α spo1Δ:HIS3 SPO1-6myc (GTY157x158); right, isogenic a/α SPO1 untagged; M, kaleidoscope standards of molecular weight (Bio-Rad, Hercules, CA). Arrows indicate positions of the 216-, 132-, and 45-kDa markers and of Spo1-6myc (∼72 kDa). (F) Western analysis of Spo1 protein accumulation during sporulation, quantified using Kodak 1D 2.0 software. Protein accumulation is shown as percentage of maximum level and sporulation landmarks as percentage of cells completing the first (MI) or both (MII) nuclear divisions and spore formation (Asci).
Media, growth, sporulation, and sporulation landmark assays:
Media:
E. coli growth media (LB and SOC) are described in Maniatis et al. (1982). The yeast media YPDA, YPA, synthetic complete (SC) for growth, and SPII and SPIII for sporulation are described in Klapholz and Esposito (1982) and Klapholz et al. (1985).
Sporulation landmarks:
Sporulation of yeast cells was carried out as reported in Klapholz et al. (1985). Briefly, cells inoculated into YPA liquid medium at ∼5 × 104 cells/ml were grown to a density of 1 × 107 cells/ml, washed twice with distilled water, and resuspended in ∼600 ml of SPII medium (supplemented with 75 μg/ml amino acids required for growth) in 4-liter flasks at 4–5 × 107 cells/ml. Sporulation cultures were incubated with vigorous aeration (250 rpm) in a New Brunswick rotary shaker at 30°. Under these conditions, the wild-type diploid strain produces >75% asci within 60 hr after transfer to sporulation medium. Recombination assays, visualization of yeast nuclei by 4′,6-diamidino-2-phenylindole (DAPI) staining, fluorescence assays for dityrosine accumulation in yeast spore walls (Briza et al. 1986), immunofluorescence staining of formaldehyde-fixed yeast cells, and preparation of thin sections for electron microscopy were performed as described elsewhere, with modifications reported earlier (Tevzadze et al. 2000).
DNA, RNA, and protein analysis:
DNA assays:
Isolation of genomic DNA for Southern blot analysis was performed as described (Hoffman and Winston 1987). Transformation of yeast utilized the one-step (Chen et al. 1992; Johnston 1994) or high-efficiency lithium acetate protocols (Gietz and Woods 1994; Gietz and Schiestl 1995). RbCl-mediated E. coli transformation of DH5α was performed as in Ano and Shoda (1992) with the following modifications in media and buffer composition. Briefly, cells were grown in 100 ml φ-broth (2% Trypton, 0.5% yeast extract, 0.4% MgSO4, and 10 mm KCl) to OD550 = 0.48, incubated on ice for 10 min, pelleted, and resuspended in 30 ml cold TJB1 (100 mm RbCl, 50 mm MnCl2, 30 mm KAc, 10 mm CaCl2, and 15% glycerol pH 5.8). The suspension was incubated on ice for 5 min, pelleted, and resuspended in 4 ml cold TJB2 (10 mm MOPS, 10 mm RbCl, 75 mm CaCl2, 15% glycerol pH 7.0). DNA was added to 100 μl cells, and the mixture was kept on ice for 30 min and transferred to 42° for 45 sec. Finally, 400 μl SOC was added and incubated with aeration at 37° for 1 hr.
RNA analysis:
Approximately 4 × 108 yeast cells were collected by centrifugation and quick frozen by submerging in liquid nitrogen. Total RNA was prepared by the glass bead/phenol protocol (Ausubel et al. 1991). Transcript levels were quantified using 15–20 μg RNA per sample by S1 nuclease protection assays and normalized against DED1 mRNA, as described (Tevzadze et al. 2000). Gels were exposed to a PhosphorImager screen and quantitated using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). All RNA probes were synthesized in SP6, T3, or T7 in vitro transcription reactions (Ausubel et al. 1991). Probes were made as follows: DED1, 0.2-kb fragment generated from AflII digestion of pGT31 (Tevzadze et al. 2000) or a 0.45-kb fragment generated from XhoI digestion of the same plasmid; SPO1, a 0.8-kb PvuII fragment from pGT29 (Tevzadze et al. 2000), which contains ∼0.6 kb of the SPO1 transcript; and SPO14, a 0.4-kb BglII–SalI fragment from pS25 (Honigberg et al. 1992).
Protein assays:
Western analyses and immunoprecipitation techniques were performed by standard procedures. Endoglycosidase H (EndoH) was obtained from ProZyme and enzymatic digestion performed as suggested by the manufacturer. After enzymatic treatment, Spo1–6myc samples were run on a 7% PAGE gel (optimal for detecting the difference between the untreated and the EndoH-treated samples).
Expression of Spo1 fragments fused to protein G was performed in BL21(DE3). Cells were grown from OD600 = 0.1–0.5 in LB plus 100 mg/ml ampicillin (for selection of the fusion plasmid) and 40 mg/ml kanamycin (for selection of the plasmid expressing T7 polymerase required for expression of the fusion). Expression was induced by addition of IPTG (Sigma, St. Louis) to a final concentration of 1 mm. After 4 hr, cells were harvested, lysed, and fusion proteins purified as described (Dames et al. 2005). Purified protein concentrations were determined by comparison to 1- to 10-μg standards on PAGE gels, and samples were diluted to 0.5 μg/ml for lipid-binding assays.
Lipid-binding assays:
Lipid-binding protein overlay assays were performed using phosphatidylinositol phosphate (PIP) strips or PIP arrays provided by Echelon, according to a protocol provided by the manufacturer (www.echelon-inc.com). PIP strips are nitrocellulose filters containing 100-pmol spots of 15 various lipids: PI and seven of its phosphorylated derivatives, phosphatidic acid (PA), lysophosphatidic acid, lysophosphocholine, phosphatidylethanolamine, phosphatidylcholine, sphingosine-1-phosphate, and phosphatidylserine. Manufacturer information indicates that binding is reproducible for all but PA. PIP arrays that allow a quantitative estimate of lipid-binding strength contain a range of seven different concentrations (100, 50, 25, 12.5, 6.25, 3.13, and 1.56 pmol) for each of the following 8 lipids (Figure 7, A and B): PtdIns, PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3. Binding to strips or arrays was assayed by ECL (Amersham, Arlington Heights, IL). Signal intensities for individual lipid spots were calculated using the NIH Image 1.62 software and normalized to the blank (PIP strips) or “no binding” (PI, arrays) spots.
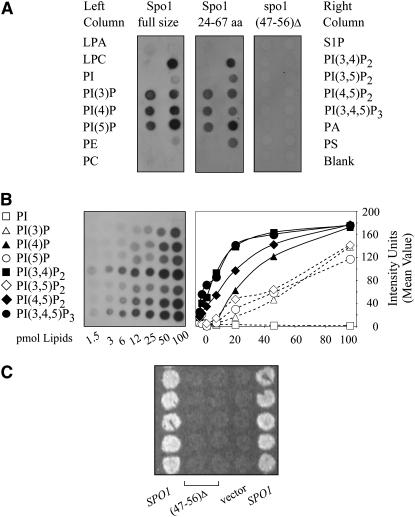
Figure 7.—
Binding of the Spo1 protein to various lipids. (A) Lipid binding using PIP strips with test lipids indicated in corresponding positions on the left and right sides of the strip. LPA, lysophophatidic acid; LPC, lysophosphocholine; PI, phosphatidylinositol; PI(3)P, phosphatidylinositol-3-monophosphate; PI(4)P, phosphatidylinositol-4-monophosphate; PI(5)P, phosphatidylinositol-5-monophosphate; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PI(3,4)P2, phosphatidylinositol-3,4-biphosphate; PI(3,5)P2, phosphatidylinositol-3,5-biphosphate; PI(4,5)P2, phosphatidylinositol-4,5-biphosphate; PI(3,4,5)P3, phosphatidylinositol-3,4,5-triphosphate; PA, phosphatidic acid; PS, phosphatidylserine; the bottom right spot does not contain any lipids. Binding is shown for full-size Spo1 (left), a 5-kDa N-terminal fragment (residues 24–67, center), and a derivative of full-size Spo1 with the lysine stretch deleted (residues 45–56, right). (B) Typical results of lipid-binding assays and their quantification for the full-size Spo1 protein using PIP arrays. Test PI and PIP lipids (abbreviated as above) are shown at the left. (C) spo1Δ complementation by wild-type SPO1, its deletion derivative with an excised lysine stretch motif, and no insert in a high-copy plasmid. Dityrosine spore fluorescence is shown for strains sporulated at 23° for 5 days.
RESULTS
The SPO1 gene product is required throughout meiosis and spore formation:
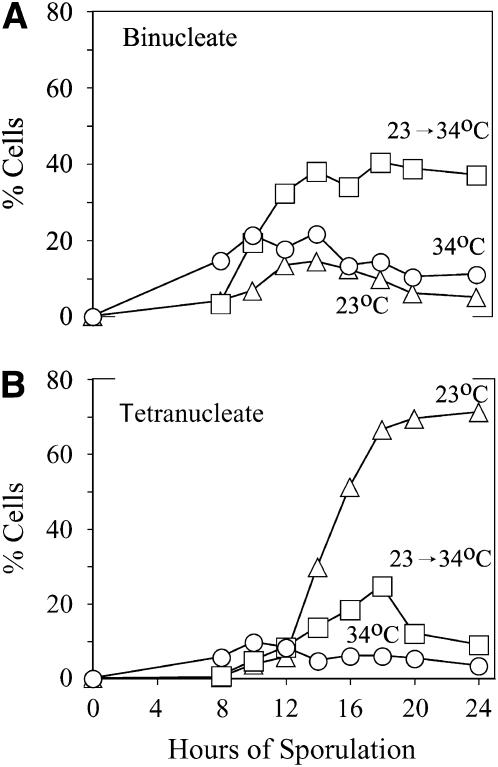
Previous studies showed spo1Δ mutants arrest at three stages of meiotic development. The major arrest occurs at MI SPB duplication, followed by two minor arrest points at MII and spore formation (Tevzadze et al. 2000). These findings are consistent with the finding that spo1-1 ts, (which sporulates normally at 23°, a permissive temperature, and exhibits a null phenotype at 34°, a restrictive one) has a temperature-sensitive period extending throughout most of sporulation (Moens 1974). They do not, however, distinguish whether the minor arrest points are a downstream consequence of the initial defect (e.g., abnormal SPBs formed at MI) or reflect a functional requirement for SPO1 at multiple stages. To determine which of these occurs, the ts allele was used in a temperature-shift experiment to monitor whether the later arrest points can occur independently of the first one. If SPO1 is required only at MI, then bypassing this arrest at a permissive temperature should allow the mutant to avoid subsequent arrest when switched to a restrictive temperature (i.e., after execution of the MI function). Conversely, if Spo1 is required at several stages during sporulation, then increased numbers of arrested cells should accumulate at the second and third arrest points after a shift to restrictive conditions. Figure 1A demonstrates that the latter is the case. Shifting cells to 34° after 8 hr of incubation at 23° (which allows completion of MI) results in a significant increase in binucleate cells that fail to progress to MII (compare >40% binucleate cells for the temperature-shifted strains to ∼20% for those sporulated at 34°). Accumulation of an increased number of tetranucleate cells likely represents an asynchronous fraction that bypassed the first two arrest points at MI and MII at 23° by the time of the shift, but not the third one at spore formation (compare >20% MII cells after temperature shift to <10% in strains maintained at 34°; Figure 1, A and B). Similarly to strains incubated constantly at 34°, the cultures shifted to 34° at 8 hr yielded no asci even after another ∼90 hr of incubation in sporulation medium (the control constantly at 23° produces 75% asci by 96 hr).
Figure 1.—
Analysis of the requirement for SPO1 at MI and later stages of sporulation. (A) Binucleate and (B) tetranucleate cells accumulate in spo1-1 ts diploid strains shifted to a restrictive temperature (34°) after 8 hr of incubation in SPII media at a permissive temperature (23°). Cells shifted from 23° to 34° (squares) are compared to controls incubated at 23° (triangles) or 34° (circles) throughout sporulation.
These and prior results lead us to conclude that Spo1 is required at multiple stages of sporulation—for SPB duplication at MI and MII and subsequently for spore development. Recent studies show that the last stage of Spo1 function occurs specifically at the transition when modified outer plaques initiate prospore membrane synthesis (our unpublished results). This is based on the fact that loss of Spo1 blocks relocalization of Don1, a meiosis-specific protein first recruited to modified SPBs and later to growing prospore membranes (Knop and Strasser 2000; Moreno-Borchart et al. 2001). Finally, the presence of spo1 mutant cells that escape arrest at the first two arrest stages (MI and MII) but not at the third (ascus formation) suggests the existence of partially redundant Spo1-independent functions/pathways promoting the meiotic nuclear divisions but not spore formation.
The Spo1 transcript and protein accumulate in a similar pattern throughout meiosis:
The regulatory region of SPO1 has an unusual and complex organization. In contrast to most yeast genes, the cis-acting regulatory sites (two URS1 elements and a UASH site) all reside within the SPO1 ORF. At least two other yeast genes have URS1 transcriptional regulatory sites within their coding regions [SPO11, a meiosis-specific gene (Atcheson 1991), and LPD1, required for amino acid catabolism (Zaman et al. 1992)]. In addition, genomewide studies of meiosis-specific transcripts that undergo splicing (Davis et al. 2000) indicate that SPO1 has a larger ORF than initially reported (Tevzadze et al. 1996), due to a previously undetected intron. Both the UASH site and the two URS1 sites, which overlap in their core sequence (GGCGG), are located in the second exon (Figure 2A).
SPO1 transcription depends upon these cis-acting elements (Figure 2B) and other factors that regulate early meiotic transcription (Figure 2, C and D). One of them, Ume6, a zinc finger protein that binds directly to the URS1 core (Strich et al. 1994; Anderson et al. 1995), interacts with a component of histone deacetylase, Sin3/Ume4 (Kadosh and Struhl 1997), to repress early genes during vegetative growth (Strich et al. 1994; Bowdish et al. 1995; Steber and Esposito 1995). Ume6 also interacts with Ime1 (Rubin-Bejerano et al. 1996), a key inducer of meiosis (Kassir et al. 1988), to both relieve Sin3-mediated mitotic repression and induce meiotic transcription (Washburn and Esposito 2001).
The data in Figure 2, B–D, show that SPO1, with its unusual internal promoter, is nonetheless regulated similarly to other early meiosis-specific genes (Atcheson et al. 1987; Buckingham et al. 1990; Vershon et al. 1992; Pittman et al. 1993; Strich et al. 1994). For example, mutations in either the URS1 elements or in UME6 cause derepressed expression in vegetative cells as well as reduced meiotic induction (Figure 2, B and D) while mutations in UASH [recognized by the Abf1 transcription factor (Prinz et al. 1995; Gailus-Durner et al. 1996)], dramatically affect meiotic induction and have only a slight, if any, defect in repression (Figure 2B). Finally, absence of the Sin3/Ume4 corepressor causes SPO1 derepression and a delay in meiotic induction (Figure 2C). Although Ime1 and Ume6 are both needed for SPO1 transcription, their own expression is unaffected by Spo1 (Tevzadze et al. 2000).
Analysis of Spo1 protein expression, using a fully functional Spo1-6myc allele, indicates that the protein begins to accumulate at ∼3 hr and starts to decline at ∼15 hr as asci form (Figure 2, E and F). The pattern closely follows transcript accumulation, which spans the time when Spo1 is required at MI, MII, and spore formation on the basis of mutant phenotype analysis. In addition to the PLB active site, which is required for its meiotic function (Tevzadze et al. 2000), Spo1 also contains another conserved motif (89SGGGYR94) defined as the “oxyanion hole,” critical for lipase activity (D. Six, personal communication; Figure 3). Intriguingly, the Spo1 oxyanion hole is encoded by the same sequence (265TCTGGCGGCGGGTACAGA282) that contains the two overlapping URS1 regulatory sites. Altering the URS1 sites and oxyanion hole by replacing 90Gly Gly Gly92 with Asp Asp Lys causes both derepressed expression and loss of spo1 complementation (monitored by light microscopy and dityrosine spore fluorescence). Since constitutive expression per se has no effect on Spo1 function in meiosis (SPO1 expressed from the TPI1 promoter shows full spo1Δ complementation; next section), we conclude that the null phenotype of the urs1 and uash spo1 mutants likely results from amino acid replacements in the oxyanion hole rather than changes in transcription. The need for both the active site as well as the oxyanion hole for Spo1 meiotic function provides additional support for the view that Spo1 acts in vivo as a meiosis-specific PLB. Furthermore, the maintenance of transcription regulatory sites in the coding region provides an unusual example of direct coupling between distinct systems controlling transcriptional regulation and protein function.
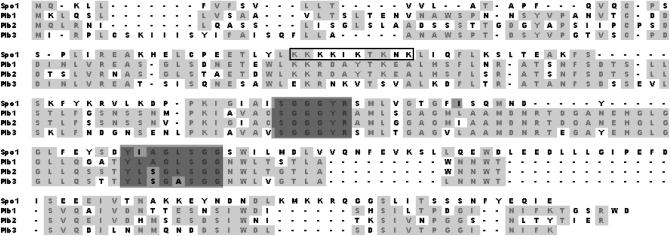
Figure 3.—
Alignment of 200 N-terminal amino acid residues for Spo1 and the Plb1, -2, and -3 enzymes of S. cerevisiae. Identical amino acid residues are shown in light shading. Conserved sequences of the oxyanion hole and the active center, both shown to be essential for PLB activities, and the Ile103 residue, replaced by Phe in Spo1-1 ts, are highlighted in dark shading. The lysine stretch motif (47KKKKIKTKNK56) in Spo1 is boxed. Note that Nte1, another protein with PLB activity (Zaccheo et al. 2004), is not shown above, since it contains neither the active site nor the oxyanion hole and has no significant similarity to Spo1 or any of the Plb proteins.
Spo1 is targeted to the ER and glycosylated similar to PLBs:
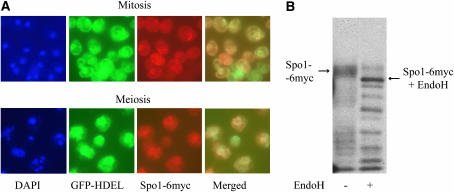
Plb1, -2, and -3 are glycosylated in the ER and subsequently localize to the plasma membrane and periplasmic space (Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999). Previously published data using partially functional Spo1-GFP on a high-copy plasmid suggested that the protein localizes throughout meiotic nuclei (Tevzadze et al. 2000). However, more extensive analysis presented here of two fully functional Spo1-6myc integrated alleles provides unambiguous evidence that Spo1 localizes around the nuclear membrane, specifically to the ER, and not throughout the nuclei as originally thought. Figure 4A (top) shows expression of Spo1-6myc from a constitutive TPI1 promoter to allow transcription in vegetative cells where ER localization is well studied. Immunostaining shows identical localization patterns for Spo1 (Texas Red) and the control ER marker, GFP-HDEL (FITC). Subsequent analysis of the same allele driven by the native SPO1 promoter confirms that Spo1 also resides in the ER of meiotic cells (Figure 4A, bottom). In both mitotic and meiotic cells, Spo1-6myc later appears to be transported to the periplasmic space, as it is detected in the supernatant following removal of the cell wall and release of periplasmic space contents (data not shown).
Figure 4.—
Localization and glycosylation of Spo1-6myc. Spheroplasts were prepared and stained with DAPI, Texas Red (for Spo1-6myc), and FITC (for GFP–HDEL) as described in materials and methods. The SPO1-6myc allele was expressed from the constitutive TPI1 promoter in vegetative cells (A, top) and during meiosis (A, bottom) from the native SPO1 promoter. (B) EndoH treatment of Spo1-6myc on a 7% PAGE gel.
Since ER-targeted proteins are generally glycosylated and Spo1 has eight putative N-glycosylation sites, Spo1 was tested for the presence of glycosyl moieties by treatment with Endo H to remove them. This caused a clear shift in mobility (Figure 4B, compare untreated and treated Spo1-6myc). These data provide further evidence that Spo1 not only requires similar motifs for function but also acts similarly to other PLB enzymes with regard to subcellular localization and post-translational modification.
High-copy GPI proteins partially suppress spo1:
Potential downstream targets, interactors and/or regulators of the likely Spo1 lipase were initially sought by screening for high-copy suppression of spo1Δ. The first suppressor recovered, CWP1 (Tevzadze et al. 2000), encodes a GPI-anchored cell wall protein (Shimoi et al. 1995; Tevzadze et al. 2000). More extensive studies below show that CWP1 acts at all three arrest points. Suppression more than doubles the level of MI and MII cells over the spo1Δ control and permits relatively efficient spore formation (∼20% asci for high-copy CWP1 compared to ∼60% for high-copy SPO1 and <0.1% for the empty vector in both null and ts mutants at 34°). Significantly, CWP1, which we previously showed is dispensable for sporulation (Tevzadze et al. 2000), plays no apparent role in redundant Spo1-independent pathways during sporulation, since the formation of bi- and tetranucleate cells is identical in spo1Δ and spo1Δ cwp1Δ strains (data not shown).
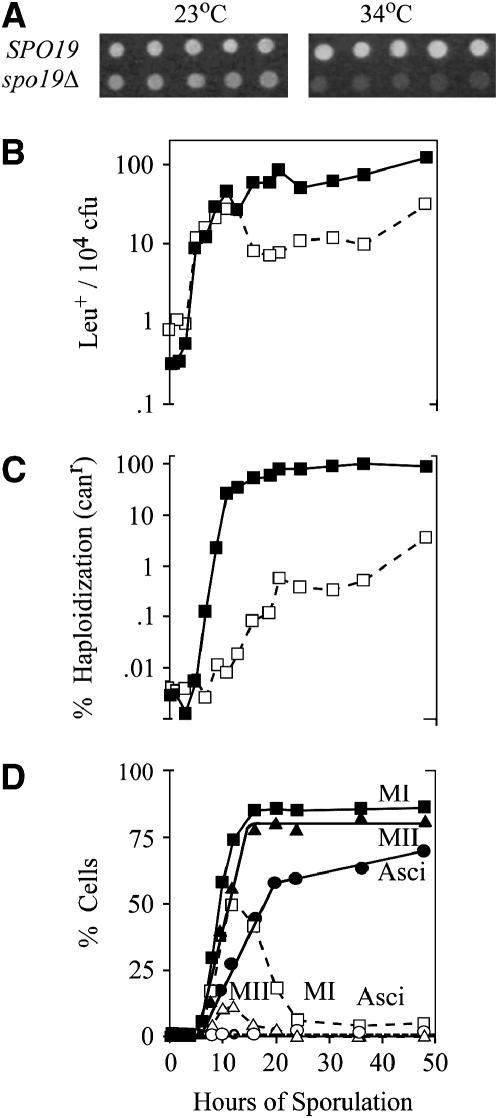
Suppression screens using the spo1-1 ts (Ile103Phe) allele in this study identified a novel meiosis-specific gene (YPL130w) along with SPO1 and CWP1. This gene, encoding another GPI protein (Caro et al. 1997) required for sporulation, is designated here SPO19. Expression of the last 40 amino acid residues of Spo19 fused to a vegetative promoter suggests that the protein localizes to the cell wall, similar to Cwp1 (Hamada et al. 1999). High-copy SPO19 has no effect on spo1Δ, but exhibits a low reproducible suppression of the ts allele (compare 3–5% asci for high-copy SPO19 to <0.1% for the ts mutant alone at the restrictive temperature, 34°). The sporulation behavior and meiosis-specific mid/late transcription of this gene suggest that it suppresses the ts phenotype only at minor arrest points (MII and/or spore formation). Interestingly, a complete deletion of SPO19 itself has a ts sporulation defect. Null mutants sporulate normally at 23° but fail to form asci at 30° and 34° in liquid media (Figure 5A), with the ts profile shifting slightly on solid media where asci form at both 23° and 30° but not 34°. The sporulation behavior of spo19Δ implies the existence of another Spo19-independent but redundant ts Spo function that acts at 23° but not at 30° and/or 34°.
Figure 5.—
Analysis of meiotic landmarks in spo19 during sporulation. Isogenic wild type and spo19Δ were monitored for (A) dityrosine at 23° and 34° (see text), (B) meiotic recombination detected by Leu+ per 104 total CFU, (C) haploidization detected by canR per total CFU, and (D) completion of MI (squares), MII (triangles), and ascus formation (circles). In B–D wild type is shown by solid symbols and solid lines, and spo19Δ is shown by open symbols and dotted lines. The assays in B–D were done at 30°.
More detailed analysis of sporulation landmarks indicates that SPO19 is dispensable for premeiotic DNA synthesis, recombination, and MI, but is required for MII and spore formation at 34° (Figure 5, B–D). As sporulation proceeds, recombinant (Leu+) and haploid (canr) cells exhibit selective death concomitant with accumulation of multinucleate cells containing fragmented nuclei. Less than 25% of cells undergo both meiotic divisions, and none form asci (Figure 5D). Thus, Spo19 is essential at the same stages of sporulation where the spo1 null exhibits its two minor arrest points. Indeed, SPO19 high-copy suppression specifically of the spo1-1 ts allele suggests that Spo19 may physically interact with the Spo1 protein at these stages.
Promoter strength and time of expression are critical for high-copy GPI-protein suppression:
The recovery of two high-copy suppressors encoding GPI proteins, one of which is dispensable for meiosis (Cwp1), raises several questions: Is the GPI attachment sequence itself sufficient for suppression? Why do other GPI proteins (e.g., Cwp2) not appear to suppress spo1? Why are the stages of suppression by Cwp1 and Spo19 different? First, the role of the GPI sequence in suppression was examined using constructs containing either (1) a truncated CWP1 allele without the GPI-attachment sequence or (2) a high-copy plasmid expressing only the GPI-attachment sequence from a CWP1 or GAL1 promoter (containing the N-terminal ER-targeting signal sequence from CWP1 inserted between the promoter and the anchor sequence for proper localization). Both constructs fail to suppress spo1. On the basis of these findings we conclude that both the GPI-attachment sequence and protein-specific sequence information are critical for suppression.
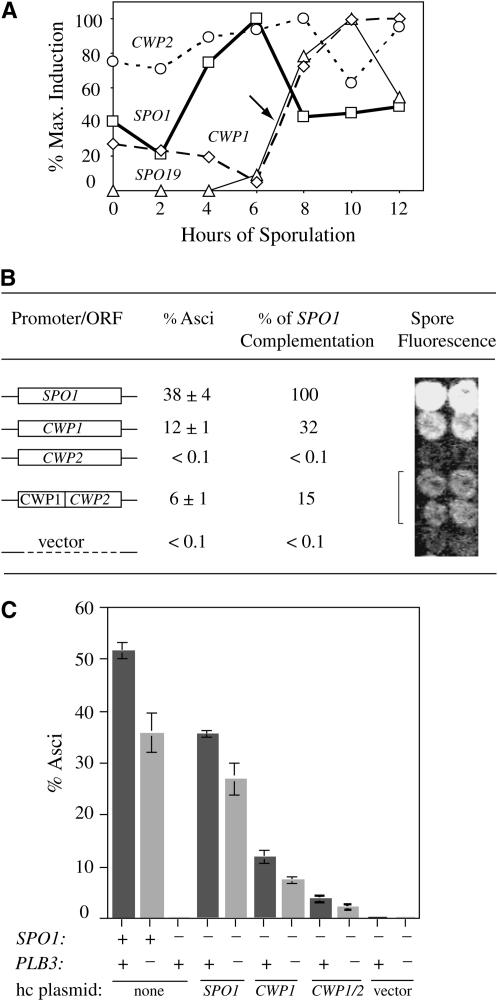
Next, promoter-swapping experiments were done to test whether the level and time of GPI-protein expression are important for suppression. For example, while both CWP1 and SPO19 are upregulated in midmeiosis, the continuous presence of the CWP1 transcript in vegetative cells and early sporulation may account for why this gene suppresses all three arrest points, in contrast to SPO19, which is specifically transcribed in meiosis over a shorter interval (Figure 6A; see also Primig et al. 2000). The lack of suppression by constitutively transcribed CWP2 might similarly be related to the absence of any meiotic regulation. Strikingly, promoter-swapping analysis shows that when the CWP2 ORF is driven by the CWP1 promoter it now suppresses spo1Δ at significantly higher levels (∼6% asci for CWP1pr-CWP2 to <0.1% for CWP2; Figure 6B). Notably, a small but reproducible increase in ascus formation is also seen in isogenic spo1-1 ts strains bearing high-copy SPO19 driven from the native or CWP1 promoter (5% vs. 7%, respectively). Even higher suppression levels (from ∼17% asci to 26%, respectively) are seen in a related spo1-1 ts strain [used in the cloning of SPO1 (Tevzadze et al. 2000)]. More efficient suppression (and complementation by SPO1) was also detected for other constructs in this strain (25% asci for CWP1, ∼1% for CWP2, 18% for CWP1pr-CWP2, and 48% for high-copy SPO1). Taken together, these results demonstrate that the efficiency of suppression by specific GPI proteins indeed depends not only on the presence of the GPI anchor and specific protein sequence, but also on expression levels, time of meiotic induction, and background modifiers.
Figure 6.—
Suppression of spo1Δ by specific GPI proteins and the effect of plb3 on suppression. (A) Comparison of the expression profiles for the SPO1, CWP1, CWP2, and SPO19 genes, based on oligo microarray analysis (Primig et al. 2000). Values are shown as percentages of maximum induction. The arrow points to similar times of upregulation for the CWP1 and SPO19 transcripts (4-fold increase for CWP1 and >1500-fold for SPO19). (B) Complementation of the spo1Δ spore formation defect by high-copy plasmids carrying SPO1, CWP1, CWP2, and a chimeric construct expressing the CWP2 ORF from the CWP1 promoter. Asci were counted for 300 cells/sample in triplicate samples for each construct after 4 days of incubation on SPIII media at 30°. (C) The effect of plb3 on spore formation and complementation/suppression of spo1Δ . Ascus production in isogenic PLB3 (dark gray), spo1Δ (clear, no asci), and plb3Δ (light gray) diploids (bars 1–3) is shown. Complementation and/or suppression of spo1Δ assayed in isogenic spo1Δ PLB3 (dark gray) and spo1Δ plb3Δ (light gray) diploids carrying high-copy vectors with SPO1 (bars 4 and 5), CWP1 (bars 6 and 7), the CWP1/2 chimera (bars 8 and 9), or no insert (bars 10 and 11) is shown.
Plb3, a lipase utilizing PI, is partially redundant to Spo1 in meiosis:
On the basis of the genetic data described above a working model was developed to explain the role of Spo1 in meiosis and the mechanism of suppression by GPI proteins. It proposes that PI utilized for the synthesis of GPI anchors also serves as a substrate of Spo1, which might act on PI/or PIPs to (a) decrease their concentration below a critical threshold and (b) produce Lyso-PI (Lyso-PIPs) and fatty acid (see discussion for details). Since PI and PI(P)s are less abundant than other intracellular phospholipids (comprising <10% of membrane lipids) and play critical roles in signaling, even small modulations in their levels can have significant consequences (Odorizzi et al. 2000; Toker 2002). For Spo1, decreasing substrate levels below a certain critical threshold as well as increasing product abundance may both serve as signals for the progression of meiosis. Accordingly, we suggest that overexpression of a GPI protein may mimic Spo1 activity, in part, by potentially titrating increased amounts of PI for anchor synthesis, thereby reducing the PI pool (and specific PIP derivatives) generating one of the potential signals needed for sporulation. How efficiently a given GPI protein does this and suppresses spo1 may depend on its sequence/conformation, rate of anchor attachment, and intracellular localization and not on whether the protein itself plays a role in meiosis (since at least one of the high-copy suppressors, Cwp1, is dispensable for the process).
These ideas were first genetically tested employing another locus, PLB3, known to alter PI levels in vivo. Among the three previously characterized PLB enzymes (expressed constitutively during growth and sporulation), Plb3 is the only one utilizing PI as a substrate (Merkel et al. 1999). The results show that PLB3 expressed from the SPO1 promoter (including internal URS1 and UAS sites, see Figure 2A) partially complements spo1Δ, resulting in a >30-fold increase in spore production (3% asci compared to <0.1% for empty vector or vector containing only the SPO1 promoter and regulatory sequences). The failure to complement at higher levels may result from localization differences and/or the specific activity of the protein. For example, Plb3, a GPI protein targeted to the ER, subsequently anchors to the plasma membrane (Caro et al. 1997; Merkel et al. 1999), while Spo1, not a GPI protein, localizes to the ER lumen and then to the periplasmic space (this study).
The absence of PLB3 alone leads to a mild but significant meiotic defect detected by reduced sporulation, reduced spo1Δ complementation by pSPO1, and a small but reproducible decrease in spo1Δ suppression (Figure 6C). These results are compatible with the idea that lack of Plb3 activity increases PI (or PIP) above a critical level so that neither Spo1 activity nor enhanced GPI-anchor synthesis (e.g., by high-copy CWP1) lowers it enough to permit wild-type sporulation. Strikingly, Spo1 and Plb3 are the only PLBs required for sporulation, with Spo1 being essential and Plb3 playing a minor role. These findings further strengthen the view that Spo1 acts as a meiosis-specific Plb3 and that PI- (or PIP)-specific phospholipases are important for meiotic progression.
At present we favor the model described above as the simplest explanation of the data. However, it should be emphasized that the role of Spo1 may be more complex (see discussion). In addition to functioning as a lipase (using PI and/or PIPs or other phospholipids as substrates), its activity on different substrates could be stimulated by PIPs and/or it could also act as a PIP-transfer protein in a manner similar to Spo14, a PLD involved in signaling known to have lipase activity (enhanced by PIP binding) as well as PIP transfer function (Sciorra et al. 1999; Rudge et al. 2001). These alternatives could also potentially reduce PIP levels with similar consequences for suppression and meiotic progression as suggested above. Furthermore, Spo1, like Plb3, may also cleave PS (Merkel et al. 1999), as well as participate in a PI signaling network. To gain further insight into Spo1 function the following studies were thus undertaken.
Spo1 binds phosphatidylinositol (4)P mono- and polyphosphates via a lysine-rich motif at the N terminus:
The idea that Spo1 utilizes PI (or PIP) as its substrate (or that its activity is enhanced by PIPs) received support from an independent proteome array analysis demonstrating that Spo1 is among ∼30 proteins that bind PI(4)P and PI(4,5)P2 (Zhu et al. 2001). Below we pursue the lipid-binding behavior of Spo1 in more detail and in addition identify the domain essential for PIP binding. These studies utilized a soluble protein G–Spo1 fusion (lacking the 24 N-most terminal amino acids of Spo1) expressed in bacteria and were initially performed with lipid strips containing 15 different lipids spotted at 100-pmol concentrations and later with arrays with a wide range of PIP concentrations (“Echelon”). The strips (Figure 7A, left) show that Spo1 binds to PIPs (not unphosphorylated PI) and to PA (not Lyso-PA) and exhibits only very weak binding to PS. Thus, we conclude that both phosphorylation of the inositol ring and the presence of the sn-2 acyl chain are important for binding. The arrays (Figure 7B), which allow better discrimination of binding specificities, further demonstrate that Spo1 preferentially binds PIPs phosphorylated in the fourth position, including PI(3,4)P2 and PI(3,4,5)P3, which were previously detected only as weak binders in the study by Zhu et al. (2001).
Surprisingly, assays with purified protein fragments derived from various deletion derivatives indicated that an internal fragment (residues 78–144), containing the oxyanion hole (89SGGGYR94) and the lipase active site (117YIAGLSGG124), does not bind lipids at all. In contrast, a nearby upstream fragment lacking these two motifs (residues 24–67) retains lipid-binding properties (Figure 7A, center). This novel region of 44 amino acids at the N terminus of Spo contains a lysine cluster motif (47KKKKIKTKNK56) critical for both Spo1 lipid-binding properties and its meiotic function. A full-size protein lacking the cluster neither binds lipids nor complements spo1Δ (Figure 7, A and C), while a 5-kDa fragment containing the 47KKKKIK52 sequence (followed by 53RRIN*57, generated by PCR errors) binds at wild-type levels (not shown). Significantly, K47 and K48 are conserved in Spo1, Plb1, and Plb2, while only K48 is present in Plb3 as well (Figure 3). Recently, a mammalian protein kinase CK2, which contains a similar lysine stretch (KKKKIKR), was also found to bind PIPs likely by positively charged lysine residues electrostatically interacting with the negatively charged head group of phosphorylated inositol (Korolchuk et al. 2005).
SPO14, critical for SPB modification during meiosis, acts in the same pathway as SPO1:
Prior meiotic analysis of SPO14 demonstrates that it is required for MII spindle development and spore formation (Honigberg et al. 1992). It encodes a phospholipase PLD that cleaves PC, stimulated by binding of PI(4,5)P2 to the protein (Rose et al. 1995a,b; Sciorra et al. 1999). Electron microscopy analysis in our laboratory (G. G. Tevzadze, unpublished results) and recently reported elsewhere (Nakanishi et al. 2006), shows that it is specifically needed for MII SPB modification and separation, as well as for initiation of the prospore membrane synthesis at SPBs. Interestingly, Spo14 is also required for Sec14-independent secretory pathways during vegetative growth (Sreenivas et al. 1998) as ts mutants of SEC14, an essential gene encoding a PI transfer protein (Xie et al. 1998), yield significantly fewer revertants in a spo14 background. Several spo14 alleles were subsequently isolated that support Sec14-independent secretion, but not sporulation, identifying two distinct functions of the Spo14 PLD during vegetative growth and sporulation (Rudge et al. 2001). Given Spo14's lipase activity, PIP-binding ability, and requirement for progression through two of the same stages of sporulation as Spo1 (MII and spore formation), we inquired whether it might interact directly or indirectly with Spo1.
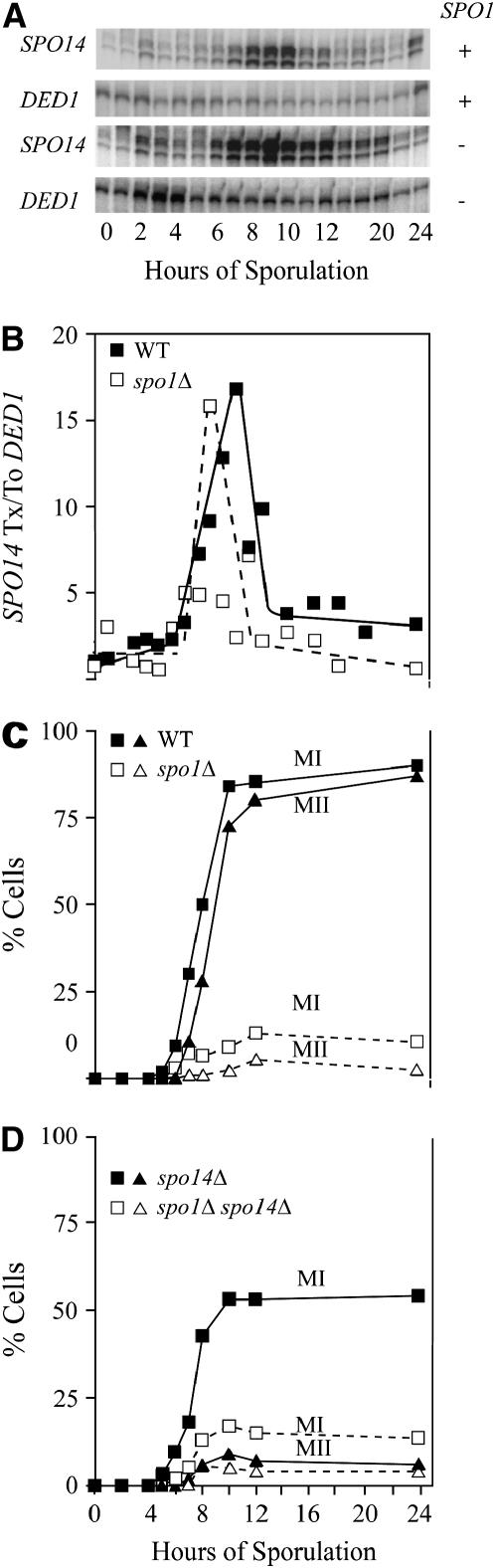
First, to test whether Spo1 might also be involved in secretory pathways we assayed sec14 revertants in strains expressing Spo1 during mitosis (under the constitutive promoter TPI1, see materials and methods) and found no effect (1.2 ± 0.6/106 colonies for sec14-1 TPI-SPO1 compared to 1.0 ± 0.6/106 for sec14-1 or sec14-1 spo1Δ). On the basis of this criterion Spo1 does not appear to play a similar role in secretion when expressed in vegetative cells. In contrast, genetic epistasis analysis suggests that SPO1 and SPO14 act in the same pathway/network during meiosis. The double mutant has a phenotype identical to spo1 with most cells being blocked at the mononucleate stage (Figure 8, C and D). Comparison of transcription profiles indicates that while SPO1 is expressed well before SPO14, epistasis does not stem from altered regulation of SPO14 as its expression is unchanged in spo1Δ and thus independent of SPO1 (Figure 8, A and B).
Figure 8.—
Analysis of interactions between Spo1 and Spo14, two phospholipase homologs required for meiosis. (A) SPO14 transcript accumulation in wild-type and spo1Δ isogenic strains relative to DED1. (B) Quantification of gel images for wild type (solid symbols, solid lines) and spo1Δ (open symbols, dotted lines). (C and D) Completion of MI (squares) and MII (triangles) for isogenic wild type (solid symbols, solid lines), spo1Δ (open symbols, dotted lines), spo14Δ (solid symbols, solid lines), and spo1Δ spo14Δ (open symbols, dotted lines).
Spo1 physically interacts with Ybr250w, a protein required for wild-type sporulation:
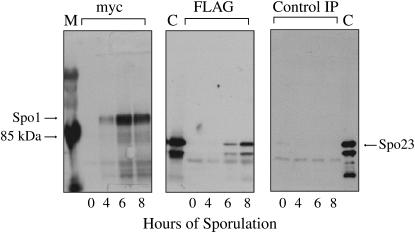
Another putative factor in the Spo1 network, an ∼60 kDa protein (Ybr-250w), was identified as a Spo1 interactor in high-throughput two-hybrid analysis (Uetz et al. 2000). Like SPO19 and SPO14, it is induced in midmeiosis (Primig et al. 2000). We confirmed that it physically interacts with Spo1 in meiosis by co-immunoprecipitation of FLAG-tagged Ybr250w with Spo1-6myc (Figure 9). Further analysis of a null revealed that it has a moderate sporulation defect (59% vs. 74% asci at 30° and 57% vs. 75% asci at 34° for isogenic ybr250wΔ and wild-type strains sporulating in liquid media for >90 hr). Given its sporulation phenotype we have designated this gene SPO23. Interestingly, SPO23/YBR250w was recently identified as one of the loci affecting expression of PIS1 (Gardocki et al. 2005), a gene encoding PI synthase essential for PI biosynthesis (Nikawa et al. 1987), implying that it is involved in regulation of lipid signaling. The precise role of Spo23 in meiosis [e.g., whether it acts as a cofactor for Spo1 lipase activity or facilitates interaction with potential substrate PI(P) molecules] remains to be determined.
Figure 9.—
Demonstration of physical interaction between Spo1 and Spo23. An a/α Spo1-6myc Spo23-FLAG strain, sampled at 0, 4, 6, and 8 hr of sporulation, was immunoprecipitated (IP) with anti-myc antibodies. Equal amounts of immunoprecipitates on 7% PAGE were probed with anti-myc (left) or anti-FLAG (center) antibodies. M, marker. C, control sample: Spo23-FLAG from 8 hr of sporulation immunoprecipitated with anti-FLAG. Control IPs (right) of an a/α Spo1 untagged, Spo23-FLAG strain did not yield detectable amounts of Spo23-FLAG (right). The positive control (C) on the right is the 8-hr sample from a/α Spo1 untagged, Spo23-FLAG immunoprecipitated and probed with anti-FLAG antibody.
DISCUSSION
This article presents a comprehensive analysis of the regulation and role of SPO1 in meiosis and spore development. Our results demonstrate the following:
The SPO1 gene product, encoding a PLB homolog that likely acts on PI(4)Ps, is required at several distinct stages of meiosis during MI, MII, and spore formation.
The Spo1 message is specifically transcribed and translated dependent on known regulators of early meiotic expression. Its cis-acting regulatory sites have an atypical location within the ORF, overlapping a site required for PLB activity and meiotic function, directly coupling regulation to function.
The protein, which persists until spore formation, is targeted to the endoplasmic reticulum (and later the periplasmic space) and is glycosylated similar to known phospholipases.
Absence of Spo1 is suppressed by high-copy expression of certain GPI proteins (depending on their sequence, promoter strength, and time of expression) and is partially complemented by PLB3 (encoding a unique PLB capable of cleaving PI), suggesting that PI and/or PIPs are Spo1 substrates.
Mono- and polyphosphate PI(4)Ps specifically bind to the N-terminal region of Spo1 (within residues 24–67), dependent on a short lysine-rich stretch, defining a novel lipid-binding domain that is essential for Spo1 function.
Epistasis analysis suggests that SPO1 acts in the same genetic pathway as SPO14, a gene encoding a phospholipase D required for MII spindle assembly, MII SPB modification, and spore formation.
Spo1 physically interacts with another protein, Spo23, which regulates expression of PSI1 (a locus essential for PI biosynthesis) and is required for wild-type levels of sporulation.
A model for the role of Spo1 in meiosis:
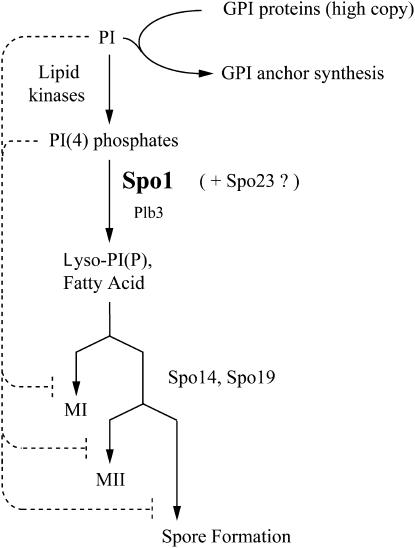
The above observations were integrated into a model for the role of the Spo1 lipase during meiosis (Figure 10). The model proposes that Spo1 functions as a meiosis-specific PLB, cleaving PI(4)Ps in a novel signal pathway regulating the timing and coordination of the MI and MII nuclear divisions and spore formation. Accordingly, decreased concentrations of PI(4)Ps below a certain threshold and increased levels of Lyso-products and fatty acids (known products of PLB and PLA2 activities) would serve as negative and positive signals, respectively, for meiotic progression. These signals may in turn directly or indirectly regulate the activities of the SPB in nucleating spindle and prospore membrane formation. The genetic and biochemical basis for this model and its implications are discussed further below.
Figure 10.—
Model for regulation of meiotic progression by the Spo1 lipase. The model proposes that a reduction in PI [and PI(4)P] levels below a critical threshold is required for progression of meiosis at MI, MII, and spore formation. This implies that at higher levels PIPs negatively regulate the process. The decrease can be achieved by Spo1 or in part by other PI-specific lipases (such as Plb3) and/or high-copy expression of GPI-anchored proteins, which utilize PI for their anchor synthesis. In this model, the accumulation of Lyso-PI and fatty acids (products of PLB activity) may also serve as positive signals for meiotic progression. Other components of this pathway identified by genetic (Spo14 and Spo19) and physical (Spo23) interaction studies are indicated at the stages where they are thought to function in meiosis (see text for details).
Two other hypotheses consistent with some of our observations are: (1) Spo1 acts as a lipase cleaving PA and/or phosphatidylserine (PS), which also bind to Spo1 and can act as second messengers, and (2) Spo1 may act as a PIP-transfer protein rather than a lipase. At present we favor the view that Spo1 acts exclusively as a lipase or as both a lipase and a PIP-transfer protein. The view that it functions only as a PIP transfer seems unlikely since the conserved lipase active site and adjacent site needed for lipase activity as well as the lipid-binding domain are essential for Spo1 meiotic function. Moreover, the idea that Spo1 acts specifically as a PIP-transfer protein makes it difficult to explain how GPI proteins suppress spo1 as there is no evidence that they regulate or function in PIP transfer. Finally, with regard to substrate specificity we think that PI(4)Ps are more likely candidates for physiological substrates as they bind to Spo1 more efficiently than PS.
Evidence that Spo1 acts as a meiotic PI-specific PLB:
Three lines of evidence support the view that Spo1 lipase activity is required in meiosis. First, Spo1 displays significant similarity to known PLB enzymes and contains two conserved motifs required for lipase activity essential for Spo1 function. Second, Plb3, a previously characterized PI-specific PLB, partially replaces Spo1 when expressed at high copy. None of the other PLBs (which act on phosphatidylethanolamine, PE, and phosphatidylcholine, PC), have the capacity to substitute for Spo1. Third, Spo1 strongly and specifically binds PI(4)P mono- and polyphosphates, well-known signaling molecules. While binding of a lipid species does not define it as a substrate (e.g., it may be a cofactor), the failure of Spo1 to bind to other conventional PLB substrates (unphosphorylated PI, PC, PE, etc.) makes it more likely that PI(4)Ps and especially PI(4,5)P2 are cleaved by Spo1. Final proof awaits direct in vitro assays of Spo1 activity, which have been hampered thus far by insufficient recovery of soluble protein. While two other strong binders, PI(3,4)P2 and PI(3,4,5)P3, have not been detected in budding yeast during vegetative growth (De Camilli et al. 1996; Vanhaesebroeck et al. 1997), it remains to be determined whether they accumulate during sporulation. Furthermore, a recent report suggests that PI(3,4,5)P3 may exist in Schizosaccharomyces pombe (Cooke 2004; Divecha and Halstead 2004; Mitra et al. 2004).
Finally, a 5-kDa N-terminal fragment was identified as necessary and sufficient for Spo1 binding to PIPs. It does not have any similarity to known lipid-binding domains, e.g., PH, PX, FYVE (Lemmon 2003), or to the PI(4,5)P2-binding region of Spo14 (Sciorra et al. 1999). It thus represents a novel PI(4)P-specific binding domain essential for Spo1 function. Within this region, a lysine cluster 47KKKKIK52 is sufficient for binding.
The evidence that PI levels coordinate and control meiotic progression:
The notion that threshold levels of PI (and its derivatives) regulate meiosis and spore development provides a plausible explanation for high-copy suppression of spo1 mutants by GPI proteins. These utilize PI in anchor synthesis, potentially reducing levels of PI (and PI(4)Ps) to partially substitute for the loss of Spo1 activity. One protein, Cwp1, apparently has no other role in meiosis and is important only when Spo1 is absent. In contrast, the other, Spo19, appears to be integrated into the normal regulatory network controlling MII and spore formation. As high-copy expression of Spo19 specifically suppresses a ts allele, it may directly interact with Spo1 to facilitate its function at these times. Since not all GPI-anchor proteins suppress spo1 and the sequence of the protein as well as timing and level of expression is critical, their localization to the cell wall in conjunction with the presence of Spo1 in the periplasmic space may be significant. Accordingly, temporal as well as spatial reduction in concentration of PI and/or PIPs may generate crucial signals.
The model described above predicts that the absence or the enhanced activity of PI(4)P-specific lipid kinases or phosphatases could also potentially substitute for Spo1 activity. On the other hand, mutant phenotypes for these functions likely will have pleiotropic effects leading not only to meiotic defects but also to impaired growth. This may be why high-copy and mutant spo1 suppressor screens did not recover such alleles. Future studies to examine the effect of mutations in specific enzymes [e.g., the Mss4 kinase required for PI(4,5)P2 synthesis and the Inp51 phosphatase, which reduces PI(4,5)P2) levels] on meiotic stages where Spo1 is required may nevertheless provide useful insights.
The evidence that SPO1 acts in a PIP-signaling pathway regulating successive stages of meiosis:
SPO1 acts at several transition points where SPBs undergo morphogenic changes during meiosis. These include SPB duplication and subsequent spindle development at MI and MII and initiation of prospore membrane synthesis at modified outer plaques. Studies of genetic interaction between SPO1 and SPO14, a gene encoding phospholipase D (Rose et al. 1995a,b), suggest that these genes act in the same pathway with SPO1 first required at MI (Tevzadze et al. 2000) and SPO14 needed later for MII, SPB modification, and spore formation (Honigberg et al. 1992; Rudge and Engebrecht 1999; Rudge et al. 2004; Riedel et al. 2005; Nakanishi et al. 2006). Recent identification and analysis of additional components of a SPO1-dependent pathway, e.g., Spo23, a physical interactor with Spo1 (this study), and Spo73, another PIP-binding protein acting downstream of the Spo1 lipase (Tevzadze et al. 2003; our unpublished results), lend further credence to the notion of a signaling network controlling meiotic progression.
Additional evidence in favor of this idea is provided by identification of several other signaling genes required for meiotic progression and spore formation. These include MAP and CDK kinases such as: (1) Smk1, a MAPK kinase required for spore morphogenesis (Krisak et al. 1994; Wagner et al. 1999); (2) Sps1, a serine–threonine kinase required for proper localization of enzymes involved in the prospore membrane synthesis (Friesen et al. 1994; Iwamoto et al. 2005); (3) Cak1, a CDK kinase required for meiotic DNA synthesis and expression of meiosis-specific loci, e.g., IME1 (Kaldis et al. 1998; Schaber et al. 2002; McDonald et al. 2005); and (4) Mps1, a dual-specificity kinase required for mitotic and meiotic SPB duplication as well as later stages of meiosis (Straight et al. 2000). How all these functions interface with one another in this relatively simple model system is not yet fully understood. Analysis of their genetic and biochemical interaction with genes defined in the Spo1 pathway described in this study should further uncover conserved signaling mechanisms coordinating the nuclear divisions with gamete maturation in yeast and higher eukaryotes.
Acknowledgments
We are indebted to our colleagues in the University of Chicago: Godfrey Getz for expert advice on lipid biochemistry and Benjamin S. Glick for generous gifts of plasmids and valuable suggestions on the ER-localization markers. Thanks are also due to Susan Gasser (Friedrich Miescher Institute, Basel, Switzerland) for helpful discussions about this work. This research was supported by National Institutes of Health grant 1RO1-GM29182 awarded to R.E.E.
References
- Anderson, S. F., C. M. Steber, R. E. Esposito and J. E. Coleman, 1995. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 4: 1832–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano, T., and M. Shoda, 1992. Ultra-rapid transformation of Escherichia coli by an alkali cation. Biosci. Biotechnol. Biochem. 56: 1505. [DOI] [PubMed] [Google Scholar]
- Atcheson, C. L., 1991. Meiosis-specific regulation of the SPO11 gene of the yeast Saccharomyces cerevisiae. Ph.D. Thesis, University of Chicago, Chicago.
- Atcheson, C. L., B. DiDomenico, S. Frackman, R. E. Esposito and R. T. Elder, 1987. Isolation, DNA sequence, and regulation of a meiosis-specific eukaryotic recombination gene. Proc. Natl. Acad. Sci. USA 84: 8035–8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 1991. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Bowdish, K. S., H. E. Yuan and A. P. Mitchell, 1995. Positive control of yeast meiotic genes by the negative regulator UME6. Mol. Cell. Biol. 15: 2955–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briza, P., G. Winkler, H. Kalchhauser and M. Breitenbach, 1986. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J. Biol. Chem. 261: 4288–4294. [PubMed] [Google Scholar]
- Buckingham, L. E., H. T. Wang, R. T. Elder, R. M. McCarroll, M. R. Slater et al., 1990. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87: 9406–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende et al., 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13: 1477–1489. [DOI] [PubMed] [Google Scholar]
- Chen, D. C., B. C. Yang and T. T. Kuo, 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21: 83–84. [DOI] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Coluccio, A., E. Bogengruber, M. N. Conrad, M. E. Dresser, P. Briza et al., 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, F. T., 2004. Phosphoinositides: Older than we first thought? Curr. Biol. 14: R762–R764. [DOI] [PubMed] [Google Scholar]
- Dames, S. A., J. M. Mulet, K. Rathgeb-Szabo, M. N. Hall and S. Grzesiek, 2005. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 280: 20558–20564. [DOI] [PubMed] [Google Scholar]
- Davis, C. A., L. Grate, M. Spingola and M. J. Ares, 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28: 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli, P., S. D. Emr, P. S. McPherson and P. Novick, 1996. Phosphoinositides as regulators in membrane traffic. Science 271: 1533–1539. [DOI] [PubMed] [Google Scholar]
- Divecha, N., and J. N. Halstead, 2004. Of yeast and men: the evolution of PtdIns(3,4,5)P3 synthesis. EMBO Rep. 5: 865–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, R. E., 2006. Meiosis and Spore Development. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 157–192.
- Friesen, H., R. Lunz, S. Doyle and J. Segall, 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8: 2162–2175. [DOI] [PubMed] [Google Scholar]
- Fyrst, H., B. Oskouian, F. A. Kuypers and J. D. Saba, 1999. The PLB2 gene of Saccharomyces cerevisiae confers resistance to phosphatidylcholine and encodes a phospholipase B/lysophospholipase. Biochemistry 38: 5864–5871. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner, V., J. Xie, C. Chintamaneni and A. K. Vershon, 1996. Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol. Cell. Biol. 16: 2777–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardocki, M. E., M. Bakewell, D. Kamath, K. Robinson, K. Borovicka et al., 2005. Genomic analysis of PIS1 gene expression. Eukaryot. Cell 4: 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. H. Schiestl, 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5: 255–269. [Google Scholar]
- Gietz, R. D., and R. A. Woods, 1994. High efficiency transformation with lithium acetate, pp. 121–134 in Molecular Genetics of Yeast: A Practical Approach, edited by J. R. Johnston. Oxford University Press, New York.
- Hamada, K., H. Terashima, M. Arisawa, N. Yabuki and K. Kitada, 1999. Amino acid residues in the omega-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J. Bacteriol. 181: 3886–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C. S., and F. Winston, 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Honigberg, S. M., C. Conicella and R. E. Esposito, 1992. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics 130: 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa, H., 2002. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 25: 409–417. [DOI] [PubMed] [Google Scholar]
- Iwamoto, M. A., S. R. Fairclough, S. A. Rudge and J. Engebrecht, 2005. Saccharomyces cerevisiae Sps1p regulates trafficking of enzymes required for spore wall synthesis. Eukaryot. Cell 4: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S. L., and M. Winey, 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20: 1–28. [DOI] [PubMed] [Google Scholar]
- Johnston, J. R., 1994. Molecular Genetics of Yeast. Oxford University Press, New York.
- Kadosh, D., and K. Struhl, 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371. [DOI] [PubMed] [Google Scholar]
- Kaldis, P., Z. W. Pitluk, I. A. Bany, D. A. Enke, M. Wagner et al., 1998. Localization and regulation of the cdk-activating kinase (Cak1p) from budding yeast. J. Cell Sci. 111: 3585–3596. [DOI] [PubMed] [Google Scholar]
- Kassir, Y., D. Granot and G. Simchen, 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52: 853–862. [DOI] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1982. A new mapping method employing a meiotic rec-mutant of yeast. Genetics 100: 387–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz, S., C. S. Waddell and R. E. Esposito, 1985. The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110: 187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and K. Strasser, 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19: 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk, V. I., G. Cozier and G. Banting, 2005. Regulation of CK2 activity by phosphatidylinositol phosphates. J. Biol. Chem. 280: 40796–40801. [DOI] [PubMed] [Google Scholar]
- Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory et al., 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8: 2151–2161. [DOI] [PubMed] [Google Scholar]
- Lee, K. S., J. L. Patton, M. Fido, L. K. Hines, S. D. Kohlwein et al., 1994. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269: 19725–19730. [PubMed] [Google Scholar]
- Lemmon, M. A., 2003. Phosphoinositide recognition domains. Traffic 4: 201–213. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E. F. Fritsch and J. Sambrook, 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mayor, S., and H. Riezman, 2004. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell. Biol. 5: 110–120. [DOI] [PubMed] [Google Scholar]
- McDonald, C. M., K. F. Cooper and E. Winter, 2005. The Ama1-directed anaphase-promoting complex regulates the Smk1 mitogen-activated protein kinase during meiosis in yeast. Genetics 171: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel, O., M. Fido, J. A. Mayr, H. Pruger, F. Raab et al., 1999. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 274: 28121–28127. [DOI] [PubMed] [Google Scholar]
- Mitra, P., Y. Zhang, L. E. Rameh, M. P. Ivshina, D. McCollum et al., 2004. A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 166: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P. B., 1974. Modification of sporulation in yeast strains with two-spored asci (Saccharomyces, Ascomycetes). J. Cell Sci. 16: 519–527. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko and M. Knop, 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20: 6946–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, H., M. Morishita, C. L. Schwartz, A. Coluccio, J. Engebrecht et al., 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119: 1406–1415. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 12: 423–440. [DOI] [PubMed] [Google Scholar]
- Neiman, A. M., 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M., 2005. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa, J., T. Kodaki and S. Yamashita, 1987. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J. Biol. Chem. 262: 4876–4881. [PubMed] [Google Scholar]
- Odorizzi, G., M. Babst and S. D. Emr, 2000. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25: 229–235. [DOI] [PubMed] [Google Scholar]
- Pittman, D., W. Lu and R. E. Malone, 1993. Genetic and molecular analysis of REC114, an early meiotic recombination gene in yeast. Curr. Genet. 23: 295–304. [DOI] [PubMed] [Google Scholar]
- Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway et al., 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. [DOI] [PubMed] [Google Scholar]
- Prinz, S., F. Klein, H. Auer, D. Schweizer and M. Primig, 1995. A DNA binding factor (UBF) interacts with a positive regulatory element in the promoters of genes expressed during meiosis and vegetative growth in yeast. Nucleic Acids Res. 23: 3449–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, C. G., M. Mazza, P. Maier, R. Korner and M. Knop, 2005. Differential requirement for phospholipase D/Spo14 and its novel interactor Sma1 for regulation of exocytotic vesicle fusion in yeast meiosis. J. Biol. Chem. 280: 37846–37852. [DOI] [PubMed] [Google Scholar]
- Rose, K., A. Morris, S. A. Rudge and J. Engebrecht, 1995. a PLD1: A Yeast Gene Essential for Meiosis Encodes a Phospholipase D. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rose, K., S. A. Rudge, M. A. Frohman, A. J. Morris and J. Engebrecht, 1995. b Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA 92: 12151–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese, O. W., C. A. Reinke, B. J. Bevis, A. T. Hammond, I. B. Sears et al., 2001. A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 153: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Bejerano, I., S. Mandel, K. Robzyk and Y. Kassir, 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16: 2518–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge, S. A., and J. Engebrecht, 1999. Regulation and function of PLDs in yeast. Biochim. Biophys. Acta 2: 167–174. [DOI] [PubMed] [Google Scholar]
- Rudge, S. A., T. R. Pettitt, C. Zhou, M. J. Wakelam and J. A. Engebrecht, 2001. SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 158: 1431–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge, S. A., V. A. Sciorra, M. Iwamoto, C. Zhou, T. Strahl et al., 2004. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis et al., 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra, V. A., S. A. Rudge, G. D. Prestwich, M. A. Frohman, J. Engebrecht et al., 1999. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 18: 5911–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J. D., R. T. Pickard, X. G. Chiou, J. V. Manetta, S. Kovacevic et al., 1994. Serine 228 is essential for catalytic activities of 85-kDa cytosolic phospholipase A2. J. Biol. Chem. 269: 23250–23254. [PubMed] [Google Scholar]
- Shimoi, H., Y. Iimura and T. Obata, 1995. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J. Biochem. 118: 302–311. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, G. J., H. van den Ende and F. M. Klis, 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147: 781–794. [DOI] [PubMed] [Google Scholar]
- Sreenivas, A., J. L. Patton-Vogt, V. Bruno, P. Griac and S. A. Henry, 1998. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J. Biol. Chem. 273: 16635–16638. [DOI] [PubMed] [Google Scholar]
- Steber, C. M., and R. E. Esposito, 1995. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. USA 92: 12490–12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, P. D., T. H. Giddings and M. Winey, 2000. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11: 3525–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich, R., R. T. Surosky, C. M. Steber, C. E. Dubois, F. Messenguy et al., 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8: 776–810. [DOI] [PubMed] [Google Scholar]
- Tevzadze, G. G., A. R. Mushegian and R. E. Esposito, 1996. The SPO1 gene product required for meiosis in yeast has a high similarity to phospholipase B enzymes. Gene 177: 253–255. [DOI] [PubMed] [Google Scholar]
- Tevzadze, G. G., H. Swift and R. E. Esposito, 2000. Spo1, a phospholipase B homolog, is required for spindle pole body duplication during meiosis in Saccharomyces cerevisiae. Chromosoma 109: 72–85. [DOI] [PubMed] [Google Scholar]
- Tevzadze, G. G., J. Pierce and R. E. Esposito, 2003. A meiotic lipid signaling system regulating nuclear divisions and spore formation in budding yeast. Yeast 20: S62. [Google Scholar]
- Toker, A., 2002. Phosphoinositides and signal transduction. Cell. Mol. Life Sci. 59: 761–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. T. A. Cagney, T. A. R. S. Mansfield, R. S. Judson et al., 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck, B., S. J. Leevers, G. Panayotou and M. D. Waterfield, 1997. Phosphoinositide 3-kinases: a conserved family of signal transducers. Trends Biochem. Sci. 22: 267–272. [DOI] [PubMed] [Google Scholar]
- Vershon, A. K., N. M. Hollingsworth and A. D. Johnson, 1992. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol. 12: 3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, M., P. Briza, M. Pierce and E. Winter, 1999. Distinct steps in yeast spore morphogenesis require distinct Smk1 MAP kinase thresholds. Genetics 151: 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn, B. K., and R. E. Esposito, 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21: 2057–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., M. Fang, M. P. Rivas, A. J. Faulkner, P. C. Sternweis et al., 1998. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA 95: 12346–12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccheo, O., D. Dinsdale, P. A. Meacock and P. Glynn, 2004. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 279: 24024–24033. [DOI] [PubMed] [Google Scholar]
- Zaman, Z., A. J. Brown and I. W. Dawes, 1992. A 3′ transcriptional enhancer within the coding sequence of a yeast gene encoding the common subunit of two multi-enzyme complexes. Mol. Microbiol. 6: 239–246. [DOI] [PubMed] [Google Scholar]
- Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor et al., 2001. Global Analysis of Protein Activities Using Proteome Chips. Science 293: 2101–2105. [DOI] [PubMed] [Google Scholar]