Abstract
Chromatin packaging directly influences gene programming as it permits only certain portions of the genome to be activated in any given developmental stage, cell, and tissue type. Histone acetyltransferases (HATs) are a key class of chromatin regulatory proteins that mediate such developmental chromatin control; however, their specific roles during multicellular development remain unclear. Here, we report the first isolation and developmental characterization of a Drosophila HAT gene (Dmel\TIP60) that is the homolog of the human HAT gene TIP60. We show that Dmel\TIP60 is differentially expressed during Drosophila development, with transcript levels significantly peaking during embryogenesis. We further demonstrate that reducing endogenous Dmel\TIP60 expression in Drosophila embryonic cells by RNAi results in cellular defects and lethality. Finally, using a GAL4-targeted RNAi system in Drosophila, we show that ubiquitous or mesoderm/muscle-specific reduction of Dmel\TIP60 expression results in lethality during fly development. Our results suggest a mechanism for HAT regulation involving developmental control of HAT expression profiles and show that Dmel\TIP60 is essential for multicellular development. Significantly, our inducible and targeted HAT knockdown system in Drosophila now provides a powerful tool for effectively studying the roles of TIP60 in specific tissues and cell types during development.
METAZOANS consist of numerous cell types, each carrying out distinct and essential roles that contribute to the growth and survival of an organism (Wolffe and Dimitrov 1993; Vermaak and Wolffe 1998; Orphanides and Reinberg 2002). Differentiation of such specialized cell lineages is achieved through the establishment and maintenance of tightly controlled gene expression profiles distinct for each cell type (Wolffe and Dimitrov 1993; Orphanides and Reinberg 2002). Such regulation in eukaryotic cells is determined in large part by the differential packaging of genes into chromatin (Wolffe and Dimitrov 1993; Vermaak and Wolffe 1998). The majority of DNA in the eukaryotic nucleus is packaged into nucleosomes, consisting of 146 bp of DNA wrapped around a histone octomer core, containing two subunits each of histones H2A, H2B, H3, and H4. Nucleosomes are, in turn, further packaged into a highly organized and compact chromatin structure through their association with nucleosomal-linking histone H1 and additional nonhistone proteins (Brand and Perrimon 1993; Wolffe and Dimitrov 1993; Fischle et al. 2003). Chromatin compaction generally makes the DNA of genes and their regulatory regions inaccessible to the transcriptional machinery and cofactor protein binding required for gene activation (Li et al. 2005). As the genome is largely maintained in this repressive chromatin state, chromatin packaging must be disrupted to accommodate protein factor binding and allow for gene activation (Wolffe and Dimitrov 1993; Roth et al. 2001; Orphanides and Reinberg 2002).
Histone-modifying enzymes termed histone acetyltransferases (HATs) are directly involved in promoting chromatin decondensation, generally resulting in positive effects on gene activation (Sterner and Berger 2000; Bottomley 2004). HATs enymatically act to catalyze the transfer of an acetyl group from acetyl-CoA to the ε-amino group of specific and conserved positively charged lysine residues within the N-terminal tails of nucleosomal histones. This modification weakens histone–DNA and neighboring nucleosomal contacts to promote chromatin disruption that, in turn, facilitates factor binding and transcriptional activation (Sterner and Berger 2000; Roth et al. 2001). A second way in which HATs regulate gene activity is through their distinct substrate preference for specific histone, lysine, and gene targets, allowing HATs to generate different acetylation patterns within the genome (Strahl and Allis 2000; Berger 2001, 2002; Fischle et al. 2003; Hake et al. 2004). Such distinct HAT-generated histone and lysine acetylation patterns, as well as additional histone modifications, have been postulated by the “histone code hypothesis” to serve as epigenetic marks that control gene expression by providing recognition sites for downstream regulatory factors (Nowak and Corces 2000; Rice and Allis 2001; Fischle et al. 2003; Bottomley 2004). Specific HATs are also capable of generating specific local or global acetylation patterns (Hebbes et al. 1994; Elefant et al. 2000a,b; Fernandez et al. 2001; Smith et al. 2001; Ho et al. 2002; Cooke et al. 2004) that influence gene expression profiles. The ability of certain HATs to acetylate nonhistone regulatory proteins adds an additional layer of complexity to their many functions (Sterner and Berger 2000). Finally, histone acetylation is a reversible process that is achieved by histone deacetylase enzymes, generally resulting in gene silencing (Alland et al. 1997). Thus, histone acetylation directly influences gene programming during development as it permits only certain portions of the genome to be activated in any given developmental stage, cell, or tissue type (Wolffe and Dimitrov 1993; Patterton and Wolffe 1996). Understanding how these differentially folded chromatin domains are created and maintained in specific cell types is of central importance to the study of biological regulation during development.
Previous reports have shown that Drosophila contains a number of human HAT homologs that belong to each of the three major HAT superfamilies: GNAT (Smith et al. 1998), MYST (Grienenberger et al. 2002), and p300/CREB-binding protein (CBP) (Akimaru et al. 1997; Ludlam et al. 2002). Their genetic analysis in Drosophila has provided essential information on the role of acetylation in a wide variety of developmental cellular processes. To gain further understanding into the developmental roles of HATs and acetylation during development, we sought to identify and characterize human HAT homologs in Drosophila (Dmel\HATs), with the reasoning that we could use such Dmel\HATs to decipher human-relevant HAT function in the multicellular Drosophila model setting (Chien et al. 2002). We chose to focus our studies on TIP60, as this HAT is representative of the MYST HAT superfamily and carries out previously described diverse roles essential for cellular function. Tip60 (tat-interactive protein, 60 kDa) was identified as part of a multimeric protein complex (Allard et al. 1999; Ikura et al. 2000; Doyon and Cote 2004) that regulates its activity in many essential cellular processes, including apoptosis (Ludlam et al. 2002; Legube et al. 2004), DNA repair (Ikura et al. 2000; Bird et al. 2002; Morrison and Shen 2005), cell cycle progression (Clarke et al. 1999), developmental cell signaling (Ceol and Horvitz 2004), ribosomal gene transcription (Reid et al. 2000; Halkidou et al. 2004), and histone variant exchange during DNA repair (Kusch et al. 2004). However, despite the importance of Tip60 in many essential cell processes, it has yet to be studied extensively in a multicellular in vivo model setting, and thus its developmental, tissue, and cell-type-specific roles remain to be explored.
Here, we report the first isolation and developmental characterization of a Drosophila HAT gene (Dmel\TIP60) that is the homolog of the human HAT gene TIP60. We present evidence that Dmel\TIP60 is differentially expressed throughout Drosophila development, with expression levels significantly peaking during embryogenesis. Using RNA interference (RNAi), we show that reducing endogenous Dmel\TIP60 expression in a Drosophila embryonic cell line results in cellular defects and lethality. Finally, we confirm this detrimental in vitro effect in vivo by using an inducible GAL4-targeted RNAi system in Drosophila and demonstrating that early ubiquitous and mesoderm-specific reduction of Dmel\TIP60 expression results in total lethality of the developing flies. Our results suggest a potential mechanism underlying HAT regulation involving developmental control of HAT expression profiles and demonstrate an essential role for Dmel\TIP60 during multicellular development.
MATERIALS AND METHODS
Identification of D. melanogaster histone acetyltransferases, isolation of cDNA clones, and DNA sequencing:
BLAST searches were carried out using the BLAST algorithm at both FlyBase (1999) and NCBI with sequences corresponding to either hTIP60 (NM_182710) or hELP3 (NM_018091). Two Drosophila expressed sequence tag (EST) clones that displayed high homology to hTIP60 and hELP3 were identified. Embryonic EST cDNA clones that matched each of these sequences (clone LD31064 for Dmel\TIP60 and RE35395 for Dmel\ELP3) were identified and then purchased from Invitrogen (Carlsbad, CA). The full open reading frames (ORFs) for each Dmel\HAT were amplified by PCR using the following primer sets. For Dmel\TIP60, the forward primer 5′-CGG CGA ATT CGC CAT CAT GAA AAT TAA CCA CAA ATA TGA G-3′ contained a EcoRI site (italics), a KOZAC sequence (underlined), and sequence corresponding to the first eight codons of Dmel\TIP60. The reverse strand primer 5′-GGT TGG ATC CTC ATC ATC ATT TGG AGC GCT TGG ACC AGT C-3′ contained a BamHI site (italics), two in-frame stop codons (underlined), and the last eight codons of Dmel\TIP60. For Dmel\ELP3, the forward primer 5′-GGC TGA ATT CGC CAT CAT GAA GGC AAA AAA GAA GTT GGG CG-3′ contained a EcoRI site (italics), a KOZAC sequence (underlined), and sequence corresponding to the first 25 bp of Dmel\ELP3. The reverse strand primer 5′-GGC CGG TCT AGA TCA TCA CTA GTT ATT TTC TTC TAT GCT CTT TGA C-3′ contained an XbaI site (italics), two in-frame stop codons (underlined), and the last 28 bp of Dmel\ELP3. PCR reactions were carried out using the Expand High Fidelity PCR system (Roche) according to the manufacturer's instructions of using 400 nm of each forward and reverse primer. The cycling parameters were 30 cycles of 95° for 2 min, 55° for 1 min, and 72° for 3 min, using Mastercycler (Eppendorf, Madison, WI). The correctly sized PCR amplification products were cloned into the TOPO pCR2.1 vector (Invitrogen) according to the manufacturer's instructions. The entire insert DNA sequence for each of these constructs was determined by the University of Pennsylvania DNA Core Sequencing Facility (Philadelphia).
Real-time PCR analysis of staged Drosophila RNA:
Total RNA was isolated from staged Canton-S. Drosophila melanogaster (12- to 24-hr embryo, first instar larvae, second instar larvae, third instar larvae, pupae, and adult flies) were treated using TRIzol (Invitrogen) and treated twice with DNA-free (Ambion, Austin, TX) to remove DNA. First-strand cDNA was prepared using the SuperScript II reverse transcriptase kit (Invitrogen) according to the manufacturer's instructions with 1 μg total RNA and 15 ng/μl of random hexamer primers (Roche). Primer sets for Dmel\ELP3 (forward primer 5′-TCC CCA TGC CGC TTG TTA GT-3′ and reverse primer 5′-CCG CCA TTG GCC ACA TAG TC-3′) amplified a 190-bp fragment. Primer sets for Dmel\TIP60 (forward primer 5′-CAC AGC GCC ACC ATT CCC TA-3′ and reverse primer 5′-CCA GAT TGT TGC CAT TCA C-3′) amplified a 202-bp fragment. All PCR reactions were carried out in triplicate in 20-μl total reaction volumes containing 0.5 units Taq (QIAGEN, Chatsworth, CA), 1 μl cDNA (from the RT reaction described above), 250 μm dNTPs (Amersham Pharmacia Biotech), 500 nm for each forward and reverse primer, and 0.25× SYBR green I dye [Molecular Probes (Eugene, OR) and Invitrogen]. The PCR was carried out in 96-well microtiter plates and the cycling conditions were 40 cycles at 95° for 45 sec, 55° for 45 sec, and 72° for 1 min with plate readings recorded after each cycle. All results were converted to real cDNA quantities by comparison to a standard curve generated with serial dilutions of either Dmel\TIP60 or Dmel\ELP3 cDNA TOPO pCR2.1 clones. All data analysis was performed using Opticon2 system software (MJ Research, Watertown, MA).
RNAi and control Dmel\TIP60 constructs:
To create the inverted-repeat Dmel\TIP60/RNAi pUAST construct, a 613-bp target RNAi sequence was amplified by PCR using primer sets specific for the Dmel\TIP60 cDNA sequence and the Dmel\TIP60 cDNA TOPO pCR2.1 clone as template. The forward primer 5′-GGA GAA TTC GCA CTG GAG TGA CCA CGC CAC AGC GCC-3′ contained an EcoRI site (italics). The reverse primer 5′-GCA TAA GAG CGG CCG CAT CTA CTG TAC TTC AGG CAG AAC TCG CAG ATG-3′ contained a NotI site (italics) and a 5-bp polylinker sequence (underlined). PCR reactions were performed as described above for Dmel\HAT cloning. The correct-size PCR-generated fragment was cloned in the sense direction into EcoRI/NotI sites in the pUAST vector under the control of the UAS promoter. This construct was designated Dmel\TIP60/pUAST.1. The same target fragment described above was next PCR amplified using the Dmel\TIP60 cDNA TOPO pCR2.1 clone as template. The forward primer 5′-GGA TCT AGA GCA CTG GAG TGA CCA CGC CAC AGC GCC-3′ contained a XbaI site (italics) and the reverse primer 5′-GCA TAA GAG CGG CCG CCT GTA CTT CAG GCA GAA CTC GCA GAT G-3′ contained a NotI site (italics). The PCR-generated fragment was cloned in an antisense orientiation into NotI and XbaI sites of the Dmel\TIP60/pUAST.1, thereby creating the inverted-repeat Dmel\TIP60/RNAi/pUAST construct. To create the sense–sense Dmel\TIP60/control construct, the same target RNAi sequence was PCR amplified with the following primers: the forward primer 5′-GCA TAA GAG CGG CCG CGC ACT GGA GTG ACC ACG CCA CAG CGC C-3′ contained a NotI site (italics) and the reverse primer 5′-GCA TCT AGA CTG TAC TTC AGG CAG AAC TCG CAG ATG-3′ contained a XbaI site (italics). The PCR-generated fragment was cloned in a sense orientiation into the NotI and XbaI sites of Dmel\TIP60/pUAST.1, creating a sense–sense Dmel\TIP60/control/pUAST construct. The PCR-generated polylinker and the common NotI restriction site that joined the two target Dmel\TIP60 repeat fragments served as the “hinge” region of the hairpin in both Dmel\TIP60/RNAi/pUAST and Dmel\TIP60/control/pUAST constructs. All cloning was carried out using standard procedures except that SURE 2 competent bacterial cells (Stratagene, La Jolla, CA) were used for all bacterial transformations to prevent recombination from occurring.
Dmel\TIP60/RNAi and control constructs for transient cell transfection were created by digesting the Dmel\TIP60/RNAi/pUAST and Dmel\TIP60/control/pUAST constructs with EcoRI and XbaI restriction enzymes, gel purifying (QIAGEN) the released fragments, and subcloning each fragment into EcoRI and XbaI restriction sites within the pAc5.1/V5-HisA vector (Invitrogen). These constructs were designated Dmel\TIP60/RNAi/pAc5.1 and Dmel\TIP60/control/pAc5.1.
Cell culture and transfection:
D.Mel-2 cells [GIBCO BRL (Gaithersburg, MD) and Invitrogen] were grown in Drosophila–serum-free media (SFM) (Invitrogen) supplemented with 90 ml/liter of 200 mm l-glutamine (GIBCO and Invitrogen). The cells were grown in a 28°, nonhumidified, ambient-air-regulated incubator (Torrey Pines Scientific) and subcultured every 3–4 days to maintain exponential growth. On day 3 postsubculture, the cells were seeded to 50–60% confluence into 35-mm plates in 2.0 ml Drosophila–SFM with l-glutamine. After an overnight incubation at 28°, the cells were incubated with the transfection mixture containing 2 μg plasmid DNA, 8 μl Cellfectin (Invitrogen), and 500 μl Drosophila–SFM without l-glutamine for 3 hr. After removal of the transfection mixture and addition of 2 ml of Drosophila–SFM with l-glutamine, each plate was incubated at 28° and observed after 24, 48, and 72 hr. As a transfection efficiency control, separate plates of cells were transfected with pAC5.1/V5-His/lacZ (Invitrogen), cells were stained using the β-Gal staining kit (Invitrogen) according to the manufacturer's instructions, and blue cells were counted to determine the transfection efficiency. All transient transfections were performed in triplicate.
Semiquantitative RT–PCR:
Total RNA either from a plate of transfected cells or from three third instar larvae progeny from a homozygous Dmel\TIP60/RNAi or control × GAL4 337 cross was isolated using TRIzol (Invitrogen) and twice treated with DNA-free (Ambion) to remove DNA. First-strand cDNA was prepared using the SuperScript II reverse transcriptase kit (Invitrogen) according to the manufacturer's instructions with 1 μg total RNA and 15 ng/μl of random hexamer primers (Roche). PCR reactions were performed in a 40-μl total volume containing 1 unit Taq (QIAGEN), 1 μl cDNA template, 250 μm dNTPs (Amersham Pharmacia Biotech), and 500 nm of each forward and reverse primer. The cycling conditions were 36 cycles of 95° for 45 sec, 55° for 45 sec, and 72° for 1 min. The forward primer (5′-TGG TAT TTC TCA CCC TAT CC-3′) and the reverse primer (5′-CAA TGA GCA GCT TGC CGT AG-3′) amplified a 427-bp fragment that corresponded to position 1407–1833 within the cDNA Dmel\TIP60 sequence.
Creation of P-element-transformed fly lines:
P-element germline transformations with pUAST constructs were performed as previously described (Elefant and Palter 1999) to create fly lines containing Dmel\TIP60/RNAi or Dmel\TIP60/control pUAST constructs. To determine on which chromosome the P-element inserted, lines heterozygous for the TM3 and TM6 balancers were mated to w1118 flies, and segregation of the w+ marker was scored: if segregation of w+ was neither with the third chromosome balancer nor with a sex chromosome, it was inferred to segregate with the second chromosome. Balancer chromosomes were subsequently crossed away by successive mating to w1118. Multiple, independent fly lines were created for each construct as the level of gene expression is dependent upon the chromosomal location of the P element, which occurs randomly.
Drosophila stocks and RNAi crosses:
For this study, the following P{pUAST}/P{pUAST} flies containing either Dmel\TIP60/RNAi or control constructs were created as described above. The GAL4 lines used were y1 w*; P{Act5C-GAL4}25FO1/CyO (donated by the Bloomington Stock Center, no. 4414; Y. Hiromi), w*;P{GawB}how24B (Brand and Perrimon 1993), and GAL4 line 337 (Elefant and Palter 1999). All crosses were performed using three males and three newly eclosed virgin females in narrow plastic vials (Applied Scientific) with yeasted Drosophila media (Jazz-Mix, Fisher Scientific) at 25°.
RESULTS
Identification and characterization of two Drosophila HAT (Dmel\HAT) genes that are homologous to the human HAT genes TIP60 and ELP3:
We first wanted to identify the human HAT homolog of MYST family member TIP60. Additionally, we also set out to identify the human HAT homolog of GNAT family member ELP3 in Drosophila so that we could compare the developmental expression profiles of two different HAT family members. Conserved sequences within the human TIP60 (hTIP60) and ELP3 (hELP3) genes were used to query the Drosophila Genome database for genomic DNA encoding homologous sequences. A single genomic clone mapping to band 4A6-B1 on the X chromosome showed significant homology to hTIP60 while a single genomic clone mapping to band 24F2 on the 2L chromosome demonstrated significant homology to hELP3. Sequences corresponding to these regions were used to conduct a BLAST search of the Drosophila EST library at FlyBase and cDNA sequences were identified that diplayed high homology to the hTIP60 sequence (listed as CG6121) and hELP3 sequence (listed as CG15433). Embryonic EST cDNA clones were identified for each Dmel\HAT (clone LD31064 for Dmel\TIP60 and RE35395 for Dmel\ELP3) and these clones were purchased and sequenced. The full sequence was determined for the ORF of each cDNA Dmel\HAT clone, designated Dmel\TIP60 and Dmel\ELP3, and aligned with its respective cDNA sequence identified in FlyBase, confirming a full ORF and a correct sequence identity for each Dmel\HAT construct.
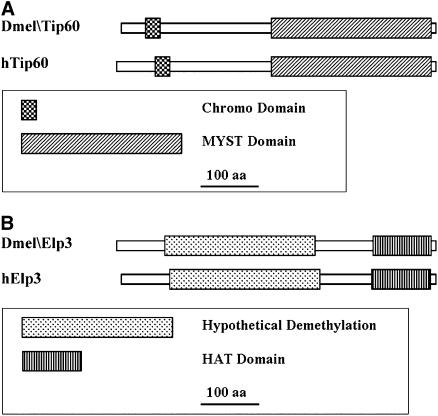
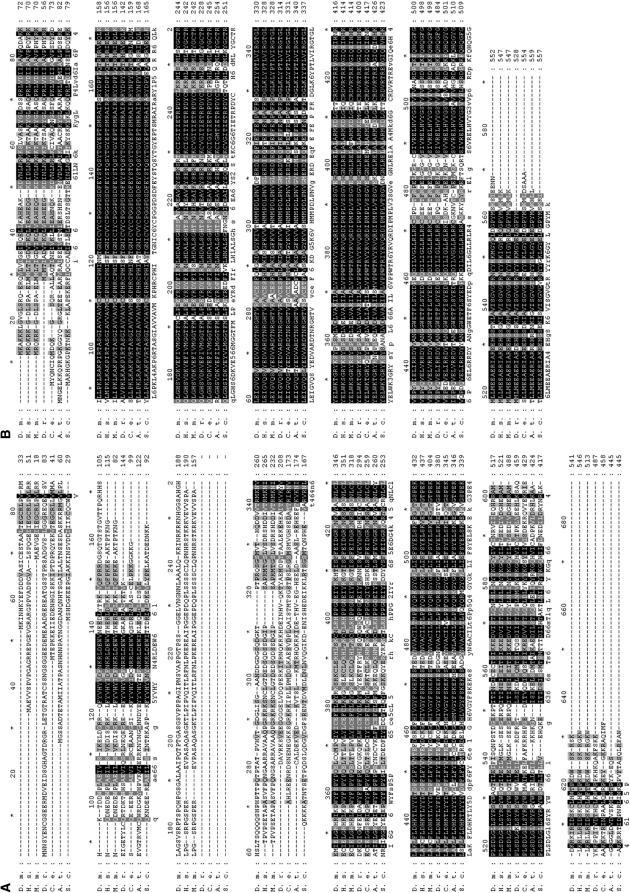
Analysis of the conceptual translation products for both Dmel\TIP60 and Dmel\ELP3 provided evidence that these Drosophila genes are homologs of the human HATs TIP60 and ELP3. First, alignments between each Dmel\HAT and its human HAT counterpart demonstrated significant homology over their entire coding sequences: Dmel\Tip60 is 58% identical/67% similar and Dmel\Elp3 is 82% identical/91% similar (Figure 1, A and B; Figure 2, A and B). Additionally, the Dmel\Tip60 transcript was found to contain an open reading frame of 1626 bp, encoding a protein of 541 aa with a predicted molecular mass of 61.2 kDa, in good agreement with the apparent molecular mass of human TIP60 (Ikura et al. 2000). The ELP3 transcript contained an ORF of 1659 bp, producing a protein of 552 aa with a predicted molecular mass of 62.8 kDa, shown to be the approximate molecular mass for the human Elp3 protein (Hawkes et al. 2002). Finally, structural protein data obtained using the conserved domain architecture retrieval tool (CDART) at NCBI revealed that the predicted protein domains specific for Dmel\Tip60 and Dmel\Elp3 and their locations within each Dmel\HAT protein are highly conserved between human and Dmel\HAT counterparts (Figure 1, A and B; Figure 2, A and B). Both Drosophila and human MYST family member Tip60 contain an N-terminal chromodomain and a C-terminal MYST domain, while both Drosophila and human GNAT family member Elp3 contain an N-terminal putative histone demethylation domain and a C-terminal HAT domain. As expected, each of these conserved domains showed significant homology to one another: for dTip60, the chromodomain is 70% identical/87% similar and the MYST domain is 80% identical/89% similar; and, for Dmel\Elp3, the HAT domain is 85% identical/93% similar while the putative histone demethylase domain is 88% identical/94% similar to its human homolog counterparts. Protein sequence analysis of a number of Dmel\Tip60 and Dmel\Elp3 homologs in a variety of different species in addition to humans, including Mus musculus, Danio rerio, Caenorhabditis elegans, Arabidopsis thaliana, and Saccharomyces cerevisiae, demonstrated that such HAT conservation for both Dmel\Tip60 and Dmel\Elp3 is evolutionarily well conserved (Figure 2, A and B). The significant sequence and structural similarity between each Dmel\HAT and its human HAT counterpart strongly indicates that these newly isolated Drosophila genes are homologs of human TIP60 and ELP3.
Figure 1.—
MYST family member Dmel\Tip60 and GNAT family member Dmel\Elp3 proteins are highly conserved with their human homolog counterparts. (A) A schematic (drawn to scale) of the conserved domains and their location within the Dmel\Tip60 and hTip60 proteins. Both proteins contain (from left to right) an N-terminal chromodomain and a C-terminal MYST functional domain. For Dmel\Tip60, the chromodomain is 70% identical/87% similar and the MYST domain is 80% identical/89% similar to hTip60. (B) Schematic (drawn to scale) of the conserved domains and their location within Dmel\Elp3 and hElp3 proteins. Both proteins contain an N-terminal putative histone demethylation domain and a C-terminal HAT domain. For Dmel\Elp3, the putative histone demethylation domain is 88% identical/94% similar and the HAT domain is 85% identical/93% similar to hElp3. (Structural domains were obtained by CDART, NCBI.)
Figure 2.—
Dmel\Tip60 and Dmel\Elp3 are evolutionarily conserved among different species. Shown are the predicted amino acid sequences for the proteins encoded by (A) Dmel\Tip60 and (B) Dmel\Elp3 and their alignment with sequences encoded by ORFs from Homo sapiens (H.s.), M. musculus (M.m.), D. rerio (D.r.), C. elegans (C.e.), A. thaliana (A.t.), and S. cerevisiae (S.c.). Interspecies homology ranges from 29 to 56% identity (D.r. to M.m.)/41 to 68% similarity (D.r. to M.m.) for Dmel\Tip60 and 70–82% identity (A.t. to H.s.)/82–92% (A.t. to H.s.) similarity for Dmel\Elp3 over their entire coding region. Solid boxes and shaded backgrounds represent identical and similar amino acids, respectively. Alignment was carried out by Genedoc.
Dmel\TIP60 and Dmel\ELP3 are differentially expressed during Drosophila development:
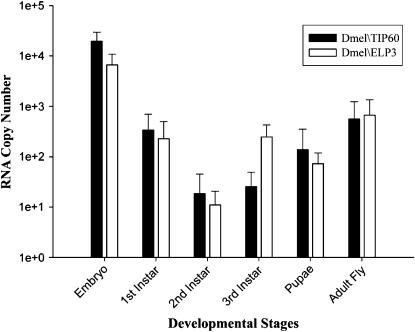
The mechanism underlying the regulation of HAT activity remains unclear. Although detailed analysis of HAT expression throughout development is limited, studies analyzing HAT expression profiles suggest that a number of HATs, including HBO1, TIP60, CBP, P/CAF, and GCN5, are controlled, at least in part, through their differential regulation in certain tissues (Xu et al. 1998, 2000; Iizuka and Stillman 1999; Stromberg et al. 1999; Lough 2002; McAllister et al. 2002). To determine whether different families of HATs might also be regulated throughout development, we examined the expression profiles of MYST family member Dmel\TIP60 and GNAT family member Dmel\ELP3 genes in all stages of Drosophila development using a real-time RT–PCR assay. RNA was isolated from staged D. melanogaster (12- to 24-hr staged embryos; first, second, and third instar larvae; pupae; adult flies) and DNaseI treated. cDNAs were generated from equal amounts of RNA for each developmental stage by RT priming with random hexamers. The RT products were then amplified in a real-time PCR assay using primer pairs corresponding to a region specific for each Dmel\HAT, and expression levels were displayed in absolute values. We found that transcript levels of both HATs significantly peaked in the embryo, sharply decreased to almost undetectable levels by the second instar larvae stage, and then gradually increased as development proceeded, reaching a second, albeit lower, peak of expression in the adult fly (Figure 3). Interestingly, although exact levels of Dmel\TIP60 and Dmel\ELP3 expression differed at each Drosophila stage tested, the trend of these levels throughout development was similar for both HATs. These data demonstrate that Dmel\TIP60 and Dmel\ELP3 are each differentially expressed throughout Drosophila development.
Figure 3.—
Dmel\TIP60 and Dmel\ELP3 are each differentially expressed during Drosophila development. We performed real-time PCR analysis of Dmel\TIP60 and Dmel\ELP3 transcript levels using stage-specific D. melanogaster cDNAs (12- to 24-hr staged embryos; first, second, and third instar larvae; pupae; adult flies) prepared by RT priming of DNase-treated RNA with random hexamers and PCR primer sets amplifying 200-bp regions specific for each dHAT. The histogram depicts RNA copy number (mean + SD) in logarithmic scale of at least three independent experiments for both Dmel\TIP60 and Dmel\ELP3 in each stage of development. SYBR-green kit and Opticon2 system (MJ Research) were used for real-time detection and data analysis. All data shown are corrected for −RT background.
Plasmid-mediated Dmel\TIP60 dsRNA production in a Drosophila embryonic cell line reduces cell viability and Dmel\TIP60 mRNA levels:
We found that levels of Dmel\TIP60 and Dmel\ELP3 expression dramatically peaked in the Drosophila embryo, supporting an important role for these Dmel\HATs during embryogenesis. Therefore, we wanted to decipher their function during early development. As no characterized Dmel\TIP60 and Dmel\ELP3 mutant alleles exist to date, we chose to silence specific endogenous HAT expression in a variety of tissues, cell types, and stages of development of choice by using an inducible GAL4-targeted RNAi-based system in Drosophila. In this RNAi/GAL4 system, expression of an inverted-repeat transgene of choice triggers double-stranded RNA (dsRNA)-mediated post-transcriptional gene silencing (Fortier and Belote 2000; Kennerdell and Carthew 2000). This method is used in conjunction with the targeted GAL4/UAS binary system (Brand and Perrimon 1993) to control expression of the inverted-repeat transgene in both a developmental and cell-type-restricted fashion.
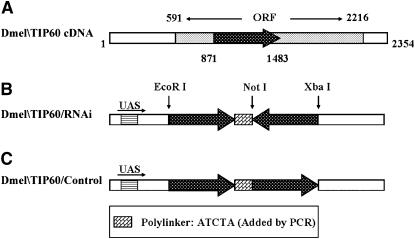
We chose to initially focus our studies on Tip60, as this HAT has been previously reported to play a wide range of biological roles essential for numerous cellular processes (Clarke et al. 1999; Ikura et al. 2000; Reid et al. 2000; Bird et al. 2002; Ceol and Horvitz 2004; Halkidou et al. 2004; Kusch et al. 2004; Legube et al. 2004). To create the Dmel\TIP60/RNAi construct, we selected a 613-bp RNAi nonconserved target sequence specific for Dmel\TIP60 (Figure 4A). BLAST searches using this sequence ensured nonredundancy within the genome. The chosen Dmel\TIP60 cDNA fragment was cloned into the inducible expression vector (pUAST) under the control of GAL4–UAS-binding sites in a sense–antisense inverted gene arrangement predicted to form a double-stranded RNA hairpin that would induce an RNAi response. This plasmid was designated the Dmel\TIP60/RNAi construct (Figure 4B). A control construct was created in which the same RNAi target sequences were cloned into a sense–sense orientation so that the control construct would not induce RNAi. This plasmid was designated the Dmel\TIP60/control construct (Figure 4C). Both the sense–antisense and sense–sense sequences in each of the constructs were separated by a short polylinker that served as the “hinge” region of the hairpin arrangement.
Figure 4.—
Structure of the pUAST Dmel\TIP60/RNAi and control constructs. (A) Schematic of the Dmel\TIP60 ORF. Solid arrow represents the location of the 613-bp RNAi nonconserved target sequence chosen for use in creating the following constructs. (B) Schematic of the Dmel\TIP60/RNAi construct. The 613-bp RNAi target cDNA sequence was amplified by PCR using the cDNA Dmel\TIP60 clone reported here as template and cloned into a sense–antisense inverted gene arrangement in the inducible expression vector (pUAST) under the control of GAL4–UAS-binding sites. A PCR-generated polylinker and the common restriction site that joins the inverted cDNA fragments separate the cloned repeats and serve as the “hinge” region of the hairpin. (C) Schematic of the Dmel\TIP60/control construct. The same RNAi cDNA target sequence was cloned into a sense–sense orientation and separated by the same short polylinker as described above.
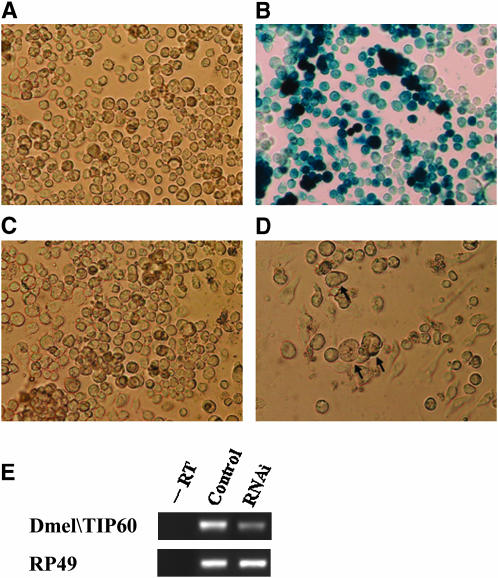
To initially test whether our Dmel\TIP60/RNAi construct would potently downregulate endogenous Dmel\TIP60 expression and result in phenotypic defects, we utilized the Drosophila embryonic D.mel-2 cell-culture-based system (Figure 5, A and B). The Dmel\TIP60/RNAi sense–antisense repeat and Dmel\TIP60/control sense–sense sequences were each subcloned into the pAc5.1/V5-HisA vector under the control of an active actin promoter. Both the Dmel\TIP60/RNAi and control constructs were transiently transfected into D.mel-2 cells and visualized using phase/contrast optics 24 hr post-transfection. We observed morphological defects in cells transfected with the Dmel\TIP60/RNAi construct. These cells were found to grow poorly, suffering ∼50–70% lethality 24 hr post-transfection (Figure 5D). Additionally, Dmel\TIP60/RNAi induction appeared to disrupt mitotic cell cycle progression, as those cells that did survive were larger than the wild-type and control cells and appeared to be arrested during cytokinesis. None of these defects were observed in cells transfected with the Dmel\TIP60/control construct (Figure 5C). These results demonstrate that Dmel\TIP60/RNAi production in a Drosophila embryonic cell line results in cellular defects and lethality, supporting an essential role for Dmel\TIP60 in early development.
Figure 5.—
The transient transfection of D.Mel-2 cells with the Dmel\TIP60/RNAi construct results in deleterious effects on cell growth and reduction of endogenous Dmel\TIP60 transcript levels. (A–D) D.Mel-2 cells visualized at ×200 magnification using phase/contrast optics. (A) Cells transiently transfected with pAc5.1/V-5-His/LacZ (unstained). (B) The same cells as in A stained with X-Gal showing transfection efficiency at 77%. (C) Cells transiently transfected with Dmel\TIP60/control construct, shown 24 hr post-transfection. (D) Cells transiently transfected with Dmel\TIP60/RNAi construct, shown 24 hr post-transfection. Arrows point to morphologically defective cells. (E) Semiquantitative RT–PCR analysis of Dmel\TIP60 and RP49 transcript levels. RNA was isolated from cells (shown above) 24 hr post-transfection. Equal amounts of RNA for each sample were subjected to cDNA preparation using RT priming with random hexamers and PCR using primer sets specific for Dmel\TIP60 that did not amplify RNAi target sequences and primer sets specific for RP49 internal control. All experiments shown were repeated at least three independent times with consistent results.
To determine whether the Dmel\TIP60/RNAi construct downregulates endogenous Dmel\TIP60, RNA was isolated from cells transfected with either the Dmel\TIP60/RNAi or the Dmel\TIP60/control construct 24 hr post-transfection and DNaseI treated. Interestingly, RNA isolated from cell plates transfected with the Dmel\TIP60/RNAi construct was found to be consistently and significantly lower in concentration than RNA isolated from cells transfected with the Dmel\TIP60/control construct (data not shown). This result is likely due to cell lethality occurring in the Dmel\TIP60/RNAi test cell lines (Figure 5D). cDNAs were generated from equal amounts of RNA for each transfection sample by RT priming with random hexamers. The RT products were amplified in a semiquantitative RT–PCR assay using primer pairs specific for each Dmel\TIP60 that did not amplify dsRNA species. The gene for the RP49 ribosomal protein was also amplified from each sample and served as an internal control. Our results revealed that endogenous Dmel\TIP60 is reduced in RNAi samples when compared to control samples, whereas RP49 expression remained unaffected. These observations indicate that our Dmel\TIP60/RNAi construct effectively and specifically inhibits endogenous Dmel\TIP60 RNA production.
Dmel\TIP60 is essential for Drosophila development:
To confirm and further explore our finding that Dmel\TIP60 is required for cell viability, we used a GAL4-targeted RNAi knockdown system to induce silencing of endogenous Dmel\TIP60 expression in the Drosophila multicellular model setting. Flies were transformed with our Dmel\TIP60/RNAi and control GAL4-inducible pUAST constructs, and three independently derived transgenic fly lines with insertions for each of the constructs were chosen for use. The insertions were homozygous viable and did not cause any observable mutant phenotypes in the absence of GAL4 induction.
On the basis of our previous findings that the actin promoter (Act5C) induced potent Dmel\TIP60 RNAi knockdown in the Drosophila cell culture line, we chose to induce Dmel\TIP60/RNAi and control transgene expression in the fly using the Act5c-Gal4 driver strain (Bloomington Stock Center no. 4414), as this actin driver expresses robust levels of GAL4 constitutively and ubiquitously early in embryogenesis (Chavous et al. 2001; Rollins et al. 2004). We found that when the Act5c-Gal4 driver was used to induce transgene expression at 25°, each of the three Dmel\TIP60/RNAi insertion lines reduced survival to 0% that of all three Dmel\TIP60/control insertion lines (Table 1). In each case, lethality for the majority of flies occurred during early pupal development, which was the latest stage that flies were able to survive. The flies that did survive until this stage showed essentially wild-type development. As an internal control, Act5c flies are hemizygous for the GAL4 driver over a CyO balancer chromosome (P{Act5c-Gal4}y/CyO y+) and thus ∼50% of flies are expected to eclose due to no GAL4 production in half of the progeny in any given cross. Thus, to determine whether a significant percentage of flies died earlier than the pupal stage, the total number of dead, noneclosed GAL4+ (y;Cy+) pupae was compared to the total number of non-RNAi-induced GAL4− (y+;Cy) flies that eclosed over a 10-day period. We found that although no Dmel\TIP60 RNAi-induced GAL4+ (y;Cy+) flies were found to eclose, the number of dead pupae was significantly lower than the number of viable GAL4− (y+;Cy) flies for one of the Dmel\TIP60/RNAi insertion lines tested. A comparison of the number of such “missing” dead pupae with the total number of eclosed GAL4− (y+;Cy) flies demonstrated that for Dmel\TIP60/RNAi/A, 24% of the Dmel\TIP60/RNAi-induced flies must have died sometime earlier than pupal development (data not shown). The variation in lethality observed between fly lines is likely due to position effects on transgene expression. Our results demonstrate that early and ubiquitous induction of Dmel\TIP60/RNAi in the fly using an actin-specific GAL4 driver results in total lethality for each of the three Dmel\TIP60/RNAi insertions tested, supporting an essential role for Dmel\TIP60 in multicellular development and the feasibility of our inducible GAL4-targeted HAT/RNAi knockdown system in Drosophila.
TABLE 1.
Ubiquitous expression of Dmel\TIP60/RNAi in three independent fly lines results in total lethality of developing flies
| Fly linesa | GAL4+(y; Cy+)b | GAL4−(y+; Cy)b |
|---|---|---|
| Dmel\TIP60/RNAi/A | 0 ± 0 | 49 ± 11 |
| Dmel\TIP60/RNAi/B | 0 ± 0 | 53 ± 12 |
| Dmel\TIP60/RNAi/C | 0 ± 0 | 57 ± 14 |
| Dmel\TIP60/control/A | 67 ± 16 | 63 ± 7 |
| Dmel\TIP60/control/B | 57 ± 0 | 59 ± 8 |
| Dmel\TIP60/control/C | 69 ± 3 | 67 ± 12 |
Three flies homozygous for either Dmel\TIP60/RNAi or Dmel\TIP60/control P-element insertions were mated to three flies homozygous for the actin GAL4 driver line Act5c-GAL4: (Dmel\TIP60/RNAi or control × P{Act5c-GAL4}/CyO,y+). For Dmel\TIP60/RNAi lines, the P-element insertion is located on the X chromosome for line A and on the second chromosome for lines B and C. For Dmel\TIP60/control lines, the P-element insertion is located on the second chromosome for line A and on the X chromosome for lines B and C.
Adult progeny were counted over a 10-day period and scored for either GAL4+(y;Cy+) or GAL4−(y+;Cy) phenotypes. All three Dmel\TIP60/RNAi lines strongly reduced viability to 0% that of the Dmel\TIP60/control lines. Lethality for the majority of flies occurred during pupal development. The results are reported as mean ±SD (n = 3).
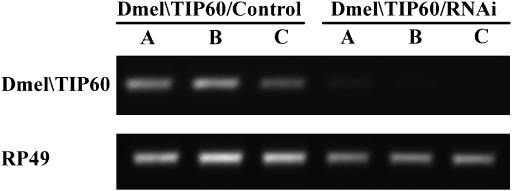
We next wanted to determine whether GAL4-induced expression of Dmel\TIP60/RNAi reduced endogenous Dmel\TIP60 transcripts. Because Act5c flies are hemizygous for the GAL4 driver, only 50% of the progeny in any given cross will induce the Dmel\TIP60/RNAi transgene, making analysis of endogenous Dmel\TIP60 downregulation using this GAL4 driver problematic. We therefore chose to induce Dmel\TIP60/RNAi and control transgenes using the ubiquitous homozygous GAL4 driver 337 (Elefant and Palter 1999). Progeny resulting from a cross between three independently derived homozygous Dmel\TIP60/RNAi or Dmel\TIP60/control fly lines and GAL4 line 337 were allowed to develop to the third instar larval stage, before lethality in the early pupal stage was shown to occur (data not shown). RNA was isolated from three third instar larvae from each of the above crosses and DNaseI treated. cDNAs were prepared from equal amounts of each RNA sample by RT priming with random hexamers. The RT products were amplified in a semiquantitative RT–PCR assay using primer pairs specific for Dmel\TIP60 that did not amplify dsRNA species. The gene for the RP49 ribosomal protein was also amplified from each sample to serve as an internal control. Our results revealed that endogenous Dmel\TIP60 transcript levels were significantly reduced in RNAi samples from each of the three independently derived Dmel\TIP60/RNAi fly lines when compared to samples obtained from each of the three independently derived Dmel\TIP60/control fly lines (Figure 6). These observations demonstrate that GAL4-induced Dmel\TIP60/RNAi expression robustly inhibits endogenous Dmel\TIP60 RNA production.
Figure 6.—
Expression of Dmel\TIP60/RNAi in three independent fly lines reduces endogenous Dmel\TIP60 levels. (A) Progeny resulting from a cross between homozygous Dmel\TIP60/RNAi (independent lines Dmel\TIP60/RNAi/A, -B, and -C) or Dmel\TIP60/control (independent lines Dmel\TIP60/control/A, -B, and -C) and ubiquitous GAL4 line 337 were allowed to develop to the third instar larval stage. RNA was isolated from three third instar larvae progeny and subjected to semiquantitative RT–PCR analysis. cDNAs were obtained from equal amounts of RNA for each sample using RT priming with random hexamers. PCR primer sets were specific for either Dmel\TIP60 that did not amplify Dmel\TIP60 RNAi target sequences or RP49. All RT–PCR experiments included negative (−RT) controls for both Dmel\TIP60 and RP49, which showed no background in all samples tested (data not shown). All experiments were repeated at least twice with consistent results. This figure shows RT–PCR analysis of one representative experiment.
Targeted expression of Dmel\TIP60/RNAi in the mesoderm and muscle cells of Drosophila results in lethal muscle mutant phenotypes:
To further test the specificity of our newly developed GAL4-targeted Dmel\TIP60/RNAi knockdown system, we wanted to determine whether targeting Dmel\TIP60/RNAi knockdown to specific tissues would result in phenotypes that were distinctive for a given particular tissue type. As our in situ analysis of Dmel\TIP60 transcripts demonstrated that Dmel\TIP60 is expressed in the muscle cells during embryogenesis (our unpublished results; data not shown; similar results in BDGP), we chose to induce Dmel\TIP60/RNAi and control transgene expression in the fly using the GAL4 line 24B (P{GawB}how24B), as this driver produces high levels of GAL4 specifically in the presumptive mesoderm and muscle cells during early embryogenesis (Brand and Perrimon 1993). Three independent fly lines containing either Dmel\TIP60/RNAi or control transgenes were crossed to the mesoderm/muscle GAL4 line 24B at 25° and the resulting phenotypes were assessed. We found that all three fly lines expressing the Dmel\TIP60 control transgene showed normal development and no observable defective phenotypes when their expression was targeted to the mesoderm/muscle cells, similar to our results for the actin-specific Act-5c and the ubiquitous 337 GAL4 drivers. However, when expression of the Dmel\TIP60/RNAi transgene was induced in the mesoderm/muscle cells, we observed a reduction in viability to 0, 40, and 29% (for lines Dmel\TIP60/RNAi/A, -B, and -C, respectively) that of the Dmel\TIP60 control lines (Table 2). Significantly, the lethal phenotypes that we observed were different from those of the Act-5c and 337 GAL4 driver lines in that, depending on the insertion line tested, the flies died at a broad range of developmental stages, beginning from early pupae to directly before fly eclosion. Importantly, the dying flies resembled those of known muscle mutants (Fyrberg et al. 1994) in that the apparent cause of lethality later in development was due to their inability to eclose from their pupal casings (data not shown). The variation in developmental lethality that we observed for different insertion lines is likely caused by position effects on transgene expression, with higher levels of Dmel\TIP60/RNAi transgene expression resulting in lethality earlier in development. Notably, the fly insertion line Dmel\TIP60/RNAi/A consistently resulted in the earliest developmental lethality of all three Dmel\TIP60/RNAi insertion lines when tested with the actin Act-5c, ubiquitous 337, and mesoderm/muscle 24B GAL4 drivers, indicating that this is the strongest expresser of our three independent Dmel\TIP60/RNAi fly lines (Tables 2). These results demonstrate the feasibility of targeting different levels of Dmel\TIP60 knockdown specifically to certain cells and tissue types and also suggest that Dmel\TIP60 is essential for proper muscle formation in the developing fly.
TABLE 2.
Mesoderm/muscle-specific expression of Dmel\TIP60/RNAi in three independent fly lines results in a range of lethal effects during fly development
| Fly linesa | Adultb | Dead pupaeb |
|---|---|---|
| Dmel\TIP60/RNAi/A | 0 ± 0 | 113 ± 26 |
| Dmel\TIP60/RNAi/B | 63 ± 15 | 74 ± 13 |
| Dmel\TIP60/RNAi/C | 46 ± 5 | 101 ± 10 |
| Dmel\TIP60/control/A | 120 ± 19 | 1 ± 1 |
| Dmel\TIP60/control/B | 179 ± 40 | 1 ± 1 |
| Dmel\TIP60/control/C | 173 ± 14 | 2 ± 1 |
Three flies homozygous for either Dmel\TIP60/RNAi or Dmel\TIP60/control P-element insertions (for P-element chromosomal locations, see Table 1) were mated to three flies homozygous for the mesoderm/muscle GAL4 driver line 24B.
Progeny were counted over a 10-day period and scored for either viable adults or dead pupae. To calculate the effect of RNAi on viability, viable progeny for each of the Dmel\TIP60/RNAi independent lines was divided by the total combined number of viable progeny for the three Dmel\TIP60/control lines. Independent insertions Dmel\TIP60/RNAi A, -B, and -C reduced viability to 0, 40, and 29%, respectively, that of the Dmel\TIP60/control lines. Lethality for the flies occurred during a broad range of developmental stages from early pupae to directly before fly eclosion. The results are reported as mean ±SD (n = 3).
DISCUSSION
The importance of histone acetylation in chromatin control and gene regulation supports a critical role for HAT function in promoting the rapidly changing gene expression profiles that drive developmental processes (Roth et al. 2001). However, the specialized roles of certain HATs in a multicellular developmental setting remains to be explored. Thus, we set out to identify human HAT family homologs in Drosophila (Dmel\HATs) to elucidate their human relevant developmental functions in the multicellular Drosophila model setting. Using homology searches of the Drosophila genome, we identified the human homologs of MYST family member TIP60 (Dmel\TIP60) and GNAT family member ELP3 (Dmel\ELP3). Our isolation and characterization of the cDNA clones encoding these genes demonstrated high conservation to their human counterparts in terms of both their amino acid sequence identity and location of conserved protein domains. Importantly, while this work was in progress Kusch et al. (2004) purified the dTip60 multiprotein complex from Drosophila embryonic S2 cells and demonstrated by mass spectrometer and sequence analysis that this complex is structurally homologous to its human counterpart and that the dTip60 protein component is encoded by the Dmel\TIP60 gene that we report here, supporting our conclusion that Dmel\TIP60 is the Drosophila homolog of human TIP60.
Our analysis of Dmel\TIP60 and Dmel\ELP3 expression levels using real-time PCR demonstrated that both Dmel\HATs are differentially expressed throughout Drosophila development. These results suggest that, in addition to being regulated by specific protein partners (Marmorstein and Roth 2001), HAT activity may also be controlled, at least in part, by their developmental regulation. In support of this idea is the observation that mice heterozygous for null alleles for each of the p300, CBP, and GCN5 HATs show less severe developmental defects than do homozygous null alleles, demonstrating that the overall dosage of HATs is critical for developmental processes (Xu et al. 2000; Roth et al. 2001). We also observed that both Dmel\TIP60 and Dmel\ELP3 expression peaked in the embryo, consistent with studies demonstrating the importance of chromatin control in early development (Patterton and Wolffe 1996). Importantly, high levels of embryonic expression are not the case for all HATs as shown by studies demonstrating that GCN5 is expressed at high levels in the mouse embryo whereas expression levels of the HAT P/CAF are virtually undetectable (Xu et al. 1998). These data, in conjunction with the HAT expression data reported here, suggest that only certain HATs may be essential for embryogenesis to proceed.
Although research on HATs in multicellular systems is still limited to date, knockout studies of p300, CBP (Tanaka et al. 1997; Roth et al. 2003), and GCN5 (Xu et al. 2000) in mice and of CBP (Akimaru et al. 1997), HBO1 (Grienenberger et al. 2002), and MOF (Smith et al. 2001) in Drosophila have revealed essential roles for these HATs during development. Significantly, the phenotypic defects that arise from such different HAT knockouts are not identical. GCN5 is essential for mouse development and formation of several mesoderm tissues while PCAF is dispensable (Xu et al. 2000), and differential roles for CBP and p300 in heart, lung, small intestine (Shikama et al. 2003), and muscle development (Roth et al. 2003) have been reported. Taken together, these studies indicate that HATs carry out specific functions required for proper multicellular development (Roth et al. 2001). Here, we show that reducing endogenous Dmel\TIP60 expression by RNAi either in all tissues or specifically in the mesoderm/muscles of the developing fly results in lethality. Our results extend prior HAT knockout studies and add Dmel\TIP60 to the growing list of HATs that carry out potentially specialized roles essential for multicellular development.
Prior studies on the yeast TIP60 homolog ESA1 demonstrated that temperature-sensitive yeast esa1 mutant cells were found to be arrested during cell division with a G2/M stage DNA content and partially depleted acetylated H4 levels, thereby linking Esa1 HAT function to cell cycle control via potential transcriptional regulatory events (Clarke et al. 1999). Consistent with these results, we observed that Dmel\TIP60 depletion in the Drosophila D.Mel-2 cell culture line resulted in a lethal phenotype reminiscent of mitotic cell cycle progression defects. Cells that did survive were larger than wild-type and control cells and appeared unable to complete cytokinesis, supporting a role for Dmel\TIP60 in metazoan embryonic cell division. We also found that either ubiquitous or mesoderm/muscle-specific depletion of Dmel\TIP60 in our GAL4-inducible HAT knockdown system resulted in lethality for all three independent Dmel\TIP60/RNAi insertion fly lines tested, with the majority of flies dying during early pupal development. Thus, as development proceeds, depletion of Dmel\TIP60 may result in the disruption of cell processes shown to require Dmel\TIP60, such as cell cycle progression (Clarke et al. 1999), apoptosis (Ikura et al. 2000; Legube et al. 2004), and DNA repair (Bird et al. 2002), as well as disruption of cell-type-specific developmental pathways, culminating in lethality caused by an accumulation of cell defects that accrue over time, all possibilities that we are currently exploring.
HATs execute acetylation profiles required for target gene regulation and thus their misregulation is linked to numerous types of cancers and developmental defects (Petrij et al. 1995; Mahlknecht et al. 2000; Steffan et al. 2001; Roelfsema et al. 2005; Close et al. 2006). The importance of TIP60 is underscored by studies demonstrating its involvement in both normal cellular processes and abnormal ones resulting in oncogenesis and developmental disorders. For example, overproduction of Tip60 in the nucleus of prostate cells is associated with androgen-resistant prostate cancer (Halkidou et al. 2003; Sapountzi et al. 2006). Tip60 is also associated with numerous disease-related proteins, including the c-MYC oncoprotein (Frank et al. 2003; Patel et al. 2004), proteins involved in hematological malignancies (Chambers et al. 2003; Nordentoft and Jorgensen 2003), and Alzheimer's-associated amyloid precursor protein (APP) (Baek et al. 2002; Kim et al. 2004). Interestingly, overproduction of the C terminus of APP induces an increase in histone acetylation that significantly enhances neurotoxicity, implicating Tip60 HAT mistargeting in Alzheimer's disease (Kim et al. 2004). Our isolation and characterization of Dmel\TIP60, in conjunction with our newly developed inducible and targeted HAT knockdown system in Drosophila, will allow us to effectively study the roles of TIP60 and other chromatin regulators in both multicellular development and epigenetic-based disorders.
Acknowledgments
The authors gratefully acknowledge Jeremy C. Lee for his invaluable scientific contributions into all aspects of this work. We also gratefully thank Karen B. Palter for generously contributing the pUAST vector and many of the fly stocks described here, as well as for sharing her cloning strategy for the construction of RNAi and control pUAST constructs. We thank Meridith Toth and Madhusmita Datta for critically reading the manuscript, Vijayalakshmi Ramakrishan for sharing her unpublished Dmel\TIP60 in situ results, Aleister Saunders and Basavaraj Hooli for their expertise in real-time PCR, and Heather Echols, Smitha Mathew, and Sandhya Manivannan for their support as lab members. Funding for this work was provided by National Institutes of Health grant HD045292-01 to F.E.
References
- Akimaru, H., Y. Chen, P. Dai, D. X. Hou, M. Nonaka et al., 1997. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386: 735–738. [DOI] [PubMed] [Google Scholar]
- Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin et al., 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387: 49–55. [DOI] [PubMed] [Google Scholar]
- Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant et al., 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, S. H., K. A. Ohgi, D. W. Rose, E. H. Koo, C. K. Glass et al., 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 110: 55–67. [DOI] [PubMed] [Google Scholar]
- Berger, S. L., 2001. An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene 20: 3007–3013. [DOI] [PubMed] [Google Scholar]
- Berger, S. L., 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12: 142–148. [DOI] [PubMed] [Google Scholar]
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon et al., 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. [DOI] [PubMed] [Google Scholar]
- Bottomley, M. J., 2004. Structures of protein domains that create or recognize histone modifications. EMBO Rep 5: 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Ceol, C. J., and H. R. Horvitz, 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6: 563–576. [DOI] [PubMed] [Google Scholar]
- Chambers, A. E., S. Banerjee, T. Chaplin, J. Dunne, S. Debernardi et al., 2003. Histone acetylation-mediated regulation of genes in leukaemic cells. Eur. J. Cancer 39: 1165–1175. [DOI] [PubMed] [Google Scholar]
- Chavous, D. A., F. R. Jackson and C. M. O'Connor, 2001. Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc. Natl. Acad. Sci. USA 98: 14814–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, S., L. T. Reiter, E. Bier and M. Gribskov, 2002. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res. 30: 149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. S., J. E. Lowell, S. J. Jacobson and L. Pillus, 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19: 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close, P., N. Hawkes, I. Cornez, C. Creppe, C. A. Lambert et al., 2006. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol. Cell 22: 521–531. [DOI] [PubMed] [Google Scholar]
- Cooke, N., Y. Ho, B. Shewchuk, A. Kimura, F. Elefant et al., 2004. Distinct epigenetic pathways activate the human growth hormone genes in pituitary and placenta, pp. 79–85 in 12th International Congress of Endocrinology, Medimond International Proceedings. Monduzzi Editore, Bologna, Italy.
- Doyon, Y., and J. Cote, 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14: 147–154. [DOI] [PubMed] [Google Scholar]
- Elefant, F., and K. B. Palter, 1999. Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol. Biol. Cell 10: 2101–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefant, F., N. E. Cooke and S. A. Liebhaber, 2000. a Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem. 275: 13827–13834. [DOI] [PubMed] [Google Scholar]
- Elefant, F., Y. Su, S. A. Liebhaber and N. E. Cooke, 2000. b Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 19: 6814–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, L. A., M. Winkler and R. Grosschedl, 2001. Matrix attachment region-dependent function of the immunoglobulin mu enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21: 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Y. Wang and C. D. Allis, 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15: 172–183. [DOI] [PubMed] [Google Scholar]
- FlyBase, 1999. The FlyBase database of the Drosophila Genome Projects and community literature. The FlyBase Consortium. Nucleic Acids Res. 27: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier, E., and J. M. Belote, 2000. Temperature-dependent gene silencing by an expressed inverted repeat in Drosophila. Genesis 26: 240–244. [DOI] [PubMed] [Google Scholar]
- Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs et al., 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg, E. A., S. I. Bernstein and K. Vijayraghavan, 1994. Basic methods for Drosophila muscle biology. Methods Cell Biol. 44: 237–258. [DOI] [PubMed] [Google Scholar]
- Grienenberger, A., B. Miotto, T. Sagnier, G. Cavalli, V. Schramke et al., 2002. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr. Biol. 12: 762–766. [DOI] [PubMed] [Google Scholar]
- Hake, S. B., A. Xiao and C. D. Allis, 2004. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br. J. Cancer 90: 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkidou, K., V. J. Gnanapragasam, P. B. Mehta, I. R. Logan, M. E. Brady et al., 2003. Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene 22: 2466–2477. [DOI] [PubMed] [Google Scholar]
- Halkidou, K., I. R. Logan, S. Cook, D. E. Neal and C. N. Robson, 2004. Putative involvement of the histone acetyltransferase Tip60 in ribosomal gene transcription. Nucleic Acids Res. 32: 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes, N. A., G. Otero, G. S. Winkler, N. Marshall, M. E. Dahmus et al., 2002. Purification and characterization of the human elongator complex. J. Biol. Chem. 277: 3047–3052. [DOI] [PubMed] [Google Scholar]
- Hebbes, T. R., A. L. Clayton, A. W. Thorne and C. Crane-Robinson, 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 13: 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., F. Elefant, N. Cooke and S. Liebhaber, 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9: 291–302. [DOI] [PubMed] [Google Scholar]
- Iizuka, M., and B. Stillman, 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274: 23027–23034. [DOI] [PubMed] [Google Scholar]
- Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang et al., 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J. R., and R. W. Carthew, 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18: 896–898. [DOI] [PubMed] [Google Scholar]
- Kim, H. S., E. M. Kim, N. J. Kim, K. A. Chang, Y. Choi et al., 2004. Inhibition of histone deacetylation enhances the neurotoxicity induced by the C-terminal fragments of amyloid precursor protein. J. Neurosci. Res. 75: 117–124. [DOI] [PubMed] [Google Scholar]
- Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser et al., 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087. [DOI] [PubMed] [Google Scholar]
- Legube, G., L. K. Linares, S. Tyteca, C. Caron, M. Scheffner et al., 2004. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 279: 44825–44833. [DOI] [PubMed] [Google Scholar]
- Li, B., C. Ruan and J. L. Workman, 2005. Histones: Should I stay or should I go? Genome Biol. 6: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough, J. W., 2002. Transient expression of TIP60 protein during early chick heart development. Dev. Dyn. 223: 419–425. [DOI] [PubMed] [Google Scholar]
- Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman et al., 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22: 3832–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlknecht, U., O. G. Ottmann and D. Hoelzer, 2000. When the band begins to play: histone acetylation caught in the crossfire of gene control. Mol. Carcinog. 27: 268–271. [DOI] [PubMed] [Google Scholar]
- Marmorstein, R., and S. Y. Roth, 2001. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 11: 155–161. [DOI] [PubMed] [Google Scholar]
- McAllister, D., X. Merlo and J. Lough, 2002. Characterization and expression of the mouse tat interactive protein 60 kD (TIP60) gene. Gene 289: 169–176. [DOI] [PubMed] [Google Scholar]
- Morrison, A. J., and X. Shen, 2005. DNA repair in the context of chromatin. Cell Cycle 4: 568–571. [PubMed] [Google Scholar]
- Nordentoft, I., and P. Jorgensen, 2003. The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor. Biochem. J. 374: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak, S. J., and V. G. Corces, 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14: 3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides, G., and D. Reinberg, 2002. A unified theory of gene expression. Cell 108: 439–451. [DOI] [PubMed] [Google Scholar]
- Patel, J. H., Y. Du, P. G. Ard, C. Phillips, B. Carella et al., 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24: 10826–10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterton, D., and A. P. Wolffe, 1996. Developmental roles for chromatin and chromosomal structure. Dev. Biol. 173: 2–13. [DOI] [PubMed] [Google Scholar]
- Petrij, F., R. H. Giles, H. G. Dauwerse, J. J. Saris, R. C. Hennekam et al., 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376: 348–351. [DOI] [PubMed] [Google Scholar]
- Reid, J. L., V. R. Iyer, P. O. Brown and K. Struhl, 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307. [DOI] [PubMed] [Google Scholar]
- Rice, J. C., and C. D. Allis, 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13: 263–273. [DOI] [PubMed] [Google Scholar]
- Roelfsema, J. H., S. J. White, Y. Ariyurek, D. Bartholdi, D. Niedrist et al., 2005. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet. 76: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, R. A., M. Korom, N. Aulner, A. Martens and D. Dorsett, 2004. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol. Cell. Biol. 24: 3100–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, J. F., N. Shikama, C. Henzen, I. Desbaillets, W. Lutz et al., 2003. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 22: 5186–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, S. Y., J. M. Denu and C. D. Allis, 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70: 81–120. [DOI] [PubMed] [Google Scholar]
- Sapountzi, V., I. R. Logan and C. N. Robson, 2006. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38: 1496–1509. [DOI] [PubMed] [Google Scholar]
- Shikama, N., W. Lutz, R. Kretzschmar, N. Sauter, J. F. Roth et al., 2003. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 22: 5175–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. R., J. M. Belote, R. L. Schiltz, X. J. Yang, P. A. Moore et al., 1998. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nucleic Acids Res. 26: 2948–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. R., C. D. Allis and J. C. Lucchesi, 2001. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 276: 31483–31486. [DOI] [PubMed] [Google Scholar]
- Steffan, J. S., L. Bodai, J. Pallos, M. Poelman, A. McCampbell et al., 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413: 739–743. [DOI] [PubMed] [Google Scholar]
- Sterner, D. E., and S. L. Berger, 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64: 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B. D., and C. D. Allis, 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Stromberg, H., S. P. Svensson and O. Hermanson, 1999. Distribution of CREB-binding protein immunoreactivity in the adult rat brain. Brain Res. 818: 510–514. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., I. Naruse, T. Maekawa, H. Masuya, T. Shiroishi et al., 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. USA 94: 10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak, D., and A. P. Wolffe, 1998. Chromatin and chromosomal controls in development. Dev. Genet. 22: 1–6. [DOI] [PubMed] [Google Scholar]
- Wolffe, A. P., and S. Dimitrov, 1993. Histone-modulated gene activity: developmental implications. Crit. Rev. Eukaryot. Gene Expr. 3: 167–191. [PubMed] [Google Scholar]
- Xu, W., D. G. Edmondson and S. Y. Roth, 1998. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol. 18: 5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer et al., 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26: 229–232. [DOI] [PubMed] [Google Scholar]