Abstract
We investigated patterns of rate asymmetry in sequence evolution among the gene pairs (ohnologs) formed by whole-genome duplication (WGD) in yeast species. By comparing three species (Saccharomyces cerevisiae, Candida glabrata, and S. castellii) that underwent WGD to a nonduplicated outgroup (Kluyveromyces lactis), and by using a synteny framework to establish orthology and paralogy relationships at each duplicated locus, we show that 56% of ohnolog pairs show significantly asymmetric protein sequence evolution. For ohnolog pairs that remain duplicated in two species there is a strong tendency for the faster-evolving copy in one species to be orthologous to the faster copy in the other species, which indicates that the evolutionary rate differences were established before speciation and hence soon after the WGD. We also present evidence that in cases where one ohnolog has been lost from the genome of a post-WGD species, the lost copy was likely to have been the faster-evolving member of the pair prior to its loss. These results suggest that a significant fraction of the retained ohnologs in yeast species underwent neofunctionalization soon after duplication.
DUPLICATION is a major motor of evolutionary innovation. Gene duplications facilitate both the acquisition of new functions (Ohno 1970) and the partitioning of old functions (Force et al. 1999; Lynch and Force 2000a). Whole-genome duplication (WGD) creates massive, albeit temporary, genomic plasticity and facilitates potentially radical biochemical innovation and species radiations (Lynch and Force 2000b; Papp et al. 2003; Scannell et al. 2006).
Duplicates are retained either because an exact duplicate provides increased dosage or because functional diversification makes their presence advantageous or even essential. It is this postduplication diversification that we examine in this study. The concept of duplication leading to functional divergence was popularized by Ohno (1970), who suggested that after duplication, one copy of a gene would retain the ancestral function and the other would be free to evolve a new function. This process is called neofunctionalization. In the past decade an alternative method of preservation of duplicates has been suggested: subfunctionalization, where both copies of the gene lose some subset of the ancestral functions, leaving two more specialized daughter genes, each of which carries out part of the ancestral function and neither of which is sufficient alone (Force et al. 1999; Lynch and Force 2000a).
Polyploidization is a particular and dramatic type of duplication process (Byrne and Blanc 2006), resulting in the duplication of all the genes in the genome and their associated regulatory elements. The simultaneous creation of these duplicates makes them ideal for studying the large-scale features of evolution after duplication. We have suggested that the duplicates arising from WGD should be called ohnologs (Wolfe 2000). Most loci quickly return to single copy after WGD, but those ohnologs that remain (typically 10–30% of the original set of loci; Byrne and Wolfe 2005; Maere et al. 2005; Paterson et al. 2006) are a major part of the ancient WGD's legacy to the organism. In this study we examine the postpolyploidy evolution of such ohnologs in multiple degenerate polyploid yeast species simultaneously. Whole-genome sequence data are available for many yeast species, including several that are descended from a common ancestor that underwent a WGD, making yeasts a model system for studying the outcome of WGD (Wolfe and Shields 1997; Dietrich et al. 2004; Kellis et al. 2004).
Theoretical considerations suggest that some form of symmetry breaking is needed for the functional diversification of duplicates (Krakauer and Nowak 1999). Previous studies have revealed asymmetric sequence divergence in 20% or more of ohnologs and non-WGD duplicates in many organisms, including yeast (Van de Peer et al. 2001; Zhang et al. 2002; Conant and Wagner 2003; Blanc and Wolfe 2004; Kellis et al. 2004). Kellis et al. (2004) found that in yeast any acceleration at a locus is typically confined to only one ohnolog and proposed that neofunctionalization had taken place at most duplicate loci showing accelerated evolution. However, their result was questioned on statistical grounds by Lynch and Katju (2004). Asymmetric sequence evolution is expected to be seen if neofunctionalization has occurred, but because subfunctionalization can also sometimes cause rate asymmetry (He and Zhang 2005) the observation of asymmetry per se is not strong evidence of neofunctionalization. The large population size of most yeast species means that on theoretical grounds subfunctionalization through the fixation of degenerative mutations by genetic drift is not expected to be a frequent mechanism of duplicate retention in yeast (Lynch and Force 2000a). Nevertheless a number of yeast gene pairs, which initially looked like candidates for classical neofunctionalization, were recently reported to be convincing examples of subfunctionalization, because homologs from an outgroup genome that diverged before the WGD compensated for the functions of both duplicates when they were knocked out (van Hoof 2005). This result has reopened the question of the extent and significance of asymmetric evolution in yeast ohnologs.
In this study we aim to identify both significantly asymmetric loci and significant trends that characterize asymmetric duplicate evolution, using a curated data set of multiple yeast genomes where relationships among orthologous and ohnolog loci are inferred using synteny relationships (Byrne and Wolfe 2005). We are primarily interested in quantifying the extent of rate asymmetry at duplicate loci, the timing of its establishment, and its relationship to neofunctionalization. Is the asymmetric fate of a pair of ohnologs established early postpolyploidization? If it is, we expect one particular member of the pair to consistently evolve faster than the other, even on nonshared phylogenetic branches. This is the first hypothesis we set out to test. Second, if an asymmetric evolutionary trajectory is established early after genome duplication we should also see a significant correlation between asymmetry on the shared and postspeciation branches, so we test for this as well. Finally, we also ask whether the pattern of asymmetric evolution we observe is characteristic of neofunctionalization. The neofunctionalization model predicts that the slower-evolving ohnologs maintain a more essential ancestral function while the faster ohnologs are free to potentially evolve a new function. Thus, if neofunctionalization is a major feature of duplicate preservation, we expect not only that asymmetric evolution is established soon after duplication, but also that the copy of the duplicate that is consistently faster evolving across a number of species is significantly more likely to be nonessential, to be uncharacterized, and even to be lost from the genome in some species.
MATERIALS AND METHODS
Genomes:
We studied the three genomes that diverged after the WGD: Saccharomyces cerevisiae (Goffeau et al. 1996), Candida glabrata (Dujon et al. 2004), and S. castellii (Cliften et al. 2003; reannotated as described in Byrne and Wolfe 2005; Cliften et al. 2006). We call these post-WGD genomes. We also include Kluyveromyces lactis (Dujon et al. 2004) in our analysis to act as an outgroup (Figure 1A). This non-WGD genome allows us to identify appropriate outgroup genes at each duplicate locus, overcoming a major difficulty faced by many previous analyses of duplicate gene evolution. We previously used the term “pre-WGD” to refer to species such as K. lactis that diverged from the S. cerevisiae lineage before WGD occurred in the latter, but we have now adopted the less confusing term “non-WGD” (Byrnes et al. 2006) for these species. We used data from the non-WGD species K. waltii (Kellis et al. 2004) and Ashbya gossypii (Dietrich et al. 2004) for some analyses. Genomic sequences and homology assignments were taken from the Yeast Gene Order Browser (YGOB) (Byrne and Wolfe 2005).
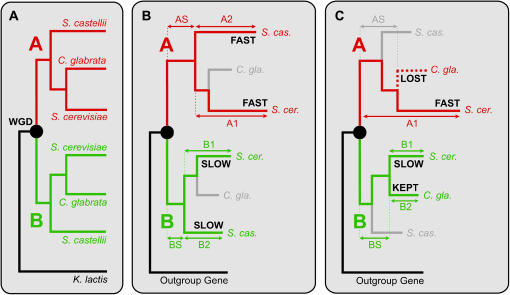
Figure 1.—
Schematic relationships among genomes and locus types. (A) Phylogenetic relationship between the two copies of a WGD duplicate (ohnolog) in the three post-WGD genomes and their ortholog in the outgroup non-WGD genome. The paralogous copies are arbitrarily labeled copy A (red) and copy B (green). The WGD event is marked by a solid circle. (B) A locus included in the Scer–Scas pairwise comparison in the 2:2 category. AS and BS are shared evolutionary branches, and A1, A2, B1, and B2 are species-specific branches. (C) A locus included in the 2:1 category of the Scer–Cgla comparison.
Pairwise species comparisons:
Phylogenetic software has difficulty recovering the correct branching relationship among the three post-WGD species, probably due to a systematic bias (Phillips et al. 2004; supplemental note S2 in Scannell et al. 2006). Since we wish to use phylogenetic signals to filter our data (see below), this is a particularly important issue. To overcome this difficulty, our analyses are based on three separate pairwise comparisons among the post-WGD species, i.e., S. cerevisiae–C. glabrata, S. cerevisiae–S. castellii, and C. glabrata–S. castellii (abbreviated as Scer–Cgla, Scer–Scas, and Cgla–Scas, respectively).
The data sets used for analysis were obtained by a series of progressively more stringent filtering processes, as described below and summarized in supplemental Table 1 at http://www.genetics.org/supplemental/.
Set 0:
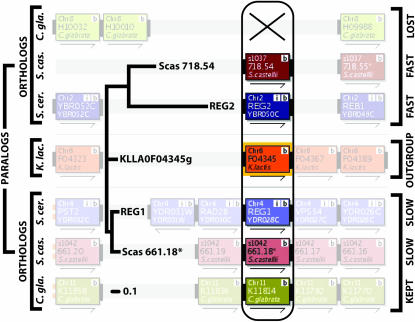
To generate our initial data set of loci the curated homology assignments from YGOB (Byrne and Wolfe 2005) were used. These contain 780 loci that have a K. lactis homolog and a pair of ohnologs present in at least one post-WGD genome. Examining these loci across the three species pairs generated 1986 data points, with the data binned into three categories: two-copy in both species (“2:2”), two-copy in the first species only (“2:1”), or two-copy in the second species only (“1:2”). We refer to this data set as set 0 (supplemental Table 1 at http://www.genetics.org/supplemental/). A locus can appear in this data set more than once. For example, in Figure 2, the K. lactis gene KLLA0F04345g is orthologous to the S. cerevisiae ohnolog pair REG1/REG2, to the S. castellii ohnolog pair Scas_661.18* and Scas_718.54, and to the C. glabrata gene CAGL0K11814g (the other member of this pair was not retained in C. glabrata). This locus was scored in the 2:2 data set for the Scer–Scas comparison, in the 2:1 data set for Scer–Cgla, and in the 1:2 data set for Cgla–Scas. To avoid any double counting of loci, the loci identified in each pairwise genome comparison are examined separately.
Figure 2.—
The REG1/REG2 locus. The YGOB (Byrne and Wolfe 2005) screenshot in the background shows the syntenic context at the locus, identifying orthologs and paralogs. The topology of the maximum-likelihood tree, from the Scer–Scas comparison in the 2:2 category, agrees perfectly with the topology derived from synteny, so this locus passes our phylogenetic filter and is retained in set 2. Furthermore, the orthologs REG2 and Scas_718.54 are the faster-evolving ohnologs in each species, making this a locus with consistent asymmetric evolution. The “X” marks the loss of the “fast” copy of the gene from C. glabrata, when it is compared to either S. cerevisiae (Scer–Cgla, 2:1) or S. castellii (Cgla–Scas, 1:2). Branch lengths show the pronounced asymmetric protein sequence evolution on both the shared and the terminal branches for this locus.
Set 1:
For each of the 1986 data points in set 0 we tested if the synteny assignment (i.e., the classification of the genes from two post-WGD species as orthologs and paralogs) by YGOB against the K. lactis genome was robust, as defined by Byrne and Wolfe (2005). To assign synteny at a locus YGOB aligns the orders of genes in paired sister regions from multiple post-WGD genomes. Robustly scored synteny at a locus means that homologous genes from the post-WGD genomes being considered are present in an unambiguously syntenic context (e.g., the two S. cerevisiae genes, the two S. castellii genes, and the single C. glabrata gene at the highlighted locus in Figure 2) and that any absent genes are absent from a clearly syntenic region of the aligned post-WGD genome (e.g., the absent C. glabrata gene marked with an “X” in Figure 2). We also checked that the scoring against two other non-WGD genomes, K. waltii and A. gossypii, did not disagree with the syntenic classification against K. lactis. These filtering steps left 1160 data points with robust scoring, and 28 of these were rejected because they caused the codeml program (see below) to fail. The remaining 1132 robust data points are referred to as set 1 (supplemental Table 1 at http://www.genetics.org/supplemental/), which contains 364 loci from the Scer–Cgla comparison, 437 loci from the Scer–Scas comparison, and 331 loci from the Cgla–Scas comparison.
Set 2:
For each data point in set 1, protein sequences for all homologous genes from the three post-WGD and the three non-WGD species were aligned using ClustalW (Chenna et al. 2003), and the gapped alignment was mapped onto those genes' nucleotide sequences. PAUP (Swofford 2003) was used to draw a maximum-likelihood (ML) tree for the locus on the basis of this nucleotide alignment. The ML trees were then pruned down to the four (2:1 and 1:2 categories) or five (2:2 category) genes of interest at that data point (i.e., the genes from the two post-WGD species and the outgroup K. lactis). Using PAUP we tested if the gene order tree (based on the YGOB scoring) and the pruned ML tree had the same topology. Figure 2 shows an example of a locus that passes this test. A total of 560 data points passed this phylogenetic filter (∼50% of the data), and we refer to these as set 2 (189 Scer–Cgla, 231 Scer–Scas, and 140 Cgla–Scas loci; supplemental Table 1 at http://www.genetics.org/supplemental/). Set 2 includes 373 data points (33% of those in set 1) where the syntenic and phylogenetic relationships agreed perfectly (row labeled “EQL” in supplemental Table 1). For the 2:2 categories an additional 187 data points produced trees with one post-WGD clade present and in agreement with YGOB, but the copies of the second ohnolog failed to recapitulate the expected species phylogeny (row “ONE” in supplemental Table 1). We consider these loci to have sufficient phylogenetic evidence to justify their use, and they are included in the set 2 data set. This filtering has a more severe effect on 2:1 and 2:1 categories than on the 2:2 categories, because with only four genes at 2:1 and 1:2 loci, phylogeny must agree either perfectly with synteny or not at all. In the 2:2 categories there were just 7 trees with the correct topology but the opposite ortholog and paralog assignments to YGOB (7 vs. 206; P = 1.14e-30 by Fisher's exact test; row “OPP” in supplemental Table 1), providing evidence that the assignments of orthology/paralogy by phylogenetic and gene order methods are in good agreement and that this is a reasonable filter for our data. Of the remaining data points in set 1 that were excluded from set 2, 439 (39%; row “GC” in supplemental Table 1) produced trees showing some evidence of gene conversion, in that both genes from one or both species formed a monophyletic group in the pruned ML tree. A further 126 trees (11%; row “OTH” in supplemental Table 1) are topologically different in other ways.
Set 3:
Using a protein alignment and the PAML (Yang 1997) program aaml with a local clock, we carried out likelihood-ratio tests (LRTs) on all loci in set 2 to test whether two distinct rate parameters for each duplicate (e.g., one rate for the S. cerevisiae gene in the A clade and another for the S. cerevisiae gene in the B clade in Figure 1C) explained the sequence data significantly better than one common rate parameter for both ohnologs. We carried out LRTs separately in each species with ohnolog pairs (i.e., in both species for 2:2 loci and in the single two-copy species for 2:1 and 1:2 loci), accounting for redundant tests at loci that are in multiple categories. To correct for multiple testing we controlled for the false discovery rate (Benjamini and Hockberg 1995). We refer to the 272 data points (49% of set 2 data) with significantly asymmetric rates of amino acid substitution in all species with ohnolog pairs (i.e., in both species for 2:2 loci and the two-copy species for 2:1 and 1:2 loci) as set 3 (95 Scer–Cgla, 97 Scer–Scas, and 80 Cgla–Scas loci; supplemental Table 1 at http://www.genetics.org/supplemental/). Across the three species, 653 ohnolog pairs were tested (267 pairs in S. cerevisiae, 168 in C. glabrata, and 218 in S. castellii) and 367 of these pairs were significantly asymmetric (56%).
For each data point in set 2 the protein sequences of the four or five genes were aligned using ClustalW (Chenna et al. 2003), and the gapped alignment was mapped onto their nucleotide sequences. Using the ML tree and the nucleotide alignment the PAML (Yang 1997) program codeml was used to estimate the rate of amino acid divergence (KA) on all branches using the free-ratio model. All sites with alignment gaps were removed. Like others (Kellis et al. 2004), we use KA rather than ω (KA/KS), because synonymous substitutions between ohnolog pairs formed by WGD are essentially saturated and because we do not expect mutation rate biases between ohnologs (Li 1997). Tests that rely on the amino acid replacement rates alone are hence more appropriate.
Computational methods and statistical analysis:
Perl scripts were used to automate and parallelize PAUP, PAML, counting, and statistical operations on the data. The R package (http://www.r-project.org) was used to carry out statistical analysis.
Yeast lethality and functional information:
Yeast viability data were downloaded in May 2004 from the Munich Information Center for Protein Sequences (MIPS) Comprehensive Yeast Genome Database (CYGD) (Guldener et al. 2005). Functional characterizations of S. cerevisiae genes were taken from the Saccharomyces Genome Database (SGD) (Christie et al. 2004) in March 2006.
RESULTS
Asymmetry is widespread and significant:
We examined rate asymmetry among ohnologs in pairwise comparisons of three paleopolyploid yeast genomes (Figure 1). To quantify the asymmetry at a locus, we defined a rate asymmetry measure, R′, for a pair of ohnologs as the maximum divided by the minimum of the rates of amino acid divergence (KA) on the terminal branches leading to those ohnologs (e.g., in Figure 1B the length of branch A1 divided by the length of branch B1 gives the R′-value for the S. cerevisiae ohnologs at the locus). In a previous study (Fares et al. 2006) we used a different measure of asymmetry (R) calculated from amino acid distances from S. cerevisiae ohnologs to C. albicans outgroup genes, but here we focus on the nonshared component of the ohnologs' evolution. For every 2:2 locus in each pairwise comparison of species an R′-value can be calculated for the ohnologs in both species, allowing comparison of the asymmetry in each since speciation.
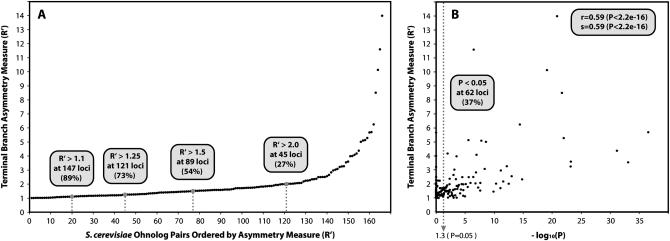
Calculating R′-values for S. cerevisiae ohnologs at all 2:2 loci in the Scer–Scas comparison that passed our filtering steps (set 2; see materials and methods) reveals that asymmetric amino acid divergence is widespread (Figure 3A). We present the Scer–Scas comparison as it is the one with the most data, but results from the Scer–Cgla and Cgla–Scas data sets are similar. All but 19 ohnologs of 166 (89%) have an amino acid distance on the terminal branch leading to one ohnolog that is >10% greater than that leading to the other (R′ > 1.1). This shows that most ohnologs are at least moderately asymmetric. More than a quarter of the data set (45 loci; 27%) are highly asymmetric with one terminal branch being more than twice the length of the other (R′ > 2).
Figure 3.—
The extent and significance of rate asymmetry. (A) Plot of the terminal branch asymmetry measure (R′) for the 166 S. cerevisiae ohnologs in the Scer–Scas data set (set 2). Loci are ordered by increasing R′ on the x-axis, with R′-values shown on the y-axis. Dashed lines show the number and percentage of loci with R′ > 1.1, 1.25, 1.5, and 2.0. (B) Correlation of R′-values for terminal branch asymmetry of S. cerevisiae ohnologs and the significance of asymmetry between those ohnologs (expressed as the negative logarithm, to base 10, of the P-value). All points to the right of the vertical dashed line are significantly asymmetric (P < 0.05) after correction for multiple testing. The correlation is highly significant for both Pearson's r and Spearman's s.
The most asymmetric locus in S. cerevisiae is REG1/REG2 (Figure 2). The slower-evolving REG1 encodes a protein three times the length of that encoded by the faster-evolving REG2 (1014 and 338 residues, respectively). The branch on the REG2 lineage prior to the S. cerevisiae/S. castellii divergence is nearly four times longer than that on the REG1 lineage, while postspeciation the ratio is 14 (in the terminology of Figure 1B, AS/BS = 0.344/0.087 and A1/B1 = 1.191/0.085). Clearly there has been very accelerated evolution on the branches leading to REG2 (Figure 2). Reg1 is similar in length to the orthologous proteins in non-WGD species, whereas Reg2 has been shortened substantially. REG1 and REG2 code for alternative regulatory subunits of protein phosphatase 1 (Glc7; Frederick and Tatchell 1996). The reason why REG2 has become much shorter and faster evolving than REG1 is not known, but there is some evidence that the two genes have diverged in function: only REG2 is activated by the oxygen-dependent transcription factor Hap1 (Ter Linde and Steensma 2002), and only REG1 plays a role in glucose repression (Frederick and Tatchell 1996; Jiang et al. 2000). Moreover, Reg2 has lost a protein domain that the amino terminus of Reg1 shares with two uncharacterized yeast proteins, Ykr075c and Yor062c (Kaniak et al. 2004).
Although R′-values give a direct measure of the asymmetry at a locus they do not tell us whether the observed asymmetry is statistically significant. For each species pair we carried out LRTs in both species at all data points to test whether two distinct rate parameters for each duplicate (e.g., one rate for the S. cerevisiae gene in the A clade and another for the S. cerevisiae gene in the B clade, and similarly for the S. castellii ohnologs, in Figure 1B) explained the sequence data significantly better than one common rate parameter for both clades. We find that the rates of amino acid substitution are significantly asymmetric in both species at 37% of loci, in the Scer–Scas comparison (P < 0.05; Figure 3B). The percentage of loci that are significantly asymmetric in both species is even higher for the 2:2 loci in the two other pairwise species comparisons (48 and 45% for the Scer–Cgla and Cgla–Scas data sets, respectively; supplemental Table 1 at http://www.genetics.org/supplemental/). Notably, the higher the value of R′ for S. cerevisiae ohnologs at a locus the more highly significant the likelihood-ratio test for those ohnologs is likely to be (Pearson's r = 0.59, Spearman's s = 0.59; P < 2.2e-16 for both; Figure 3B). Overall, across all three species, 56% of ohnolog pairs show significantly asymmetric protein sequence evolution.
Asymmetry is consistent across species:
Asymmetry is clearly widespread among yeast ohnologs within a species, but we also wish to examine whether asymmetry is consistent across post-WGD species. In other words, is the same copy the faster-evolving one in multiple post-WGD species? For each 2:2 category, and examining only loci that have significantly asymmetric rates of amino acid divergence (KA) in both species (set 3; see materials and methods), we tested whether the faster-evolving copy of the duplicate in one species is also the faster-evolving ohnolog in a second species. We use the term “consistently asymmetric” to describe loci of this type. For example, the locus in Figure 1B is consistently asymmetric because the A1 branch is longer than the B1 branch, and the A2 branch is longer than the B2 branch. Note that we exclude shared evolutionary history in the two species by comparing only values of KA on the terminal branches after the speciation event.
At 89–90% of loci the faster-evolving ohnolog in one species is the faster-evolving ohnolog in the other species too (Table 1A, “Fast (sp. 1) is fast (sp. 2)” row; P < 0.0001 in each of the three comparisons). In other words, for any species pair, the faster-evolving copy in one species is significantly more likely to be the ortholog rather than the paralog of the faster copy in the other species.
TABLE 1.
Consistency of asymmetric evolution
|
S. cerevisiae–C. glabrata
|
S. cerevisiae–S. castellii
|
C. glabrata–S. castellii
|
||||
|---|---|---|---|---|---|---|
| Locus statusa | Loci | % | Loci | % | Loci | % |
| A. | ||||||
| Fast (sp. 1) is fast (sp. 2) | 58 | 89 | 55 | 89 | 37 | 90 |
| Fast (sp. 1) is slow (sp. 2) | 7 | 11 | 7 | 11 | 4 | 10 |
| Total no. of loci | 65 | 100 | 62 | 100 | 41 | 100 |
| P = 1.06e-6 | P = 4.37e-6 | P = 7.94e-5 | ||||
| B. | ||||||
| Shared branch is fast | 52 | 90 | 45 | 82 | 33 | 89 |
| Shared branch is slow | 6 | 10 | 10 | 18 | 4 | 11 |
| Total fast (sp. 1) is fast (sp. 2) | 58 | 100 | 55 | 100 | 37 | 100 |
| P = 4.81e-6 | P = 5.60e-4 | P = 3.15e-4 | ||||
The data set used is set 3 (2:2 categories; supplemental Table 1 at http://www.genetics.org/supplemental/). Rates of amino acid divergence are examined on (A) terminal branches and (B) shared branches. P-values are from Fisher's exact two-tail tests against neutral expectation.
“Fast (sp. 1) is fast (sp. 2)” means that the same ohnolog is the faster-evolving one in both species. “Fast (sp. 1) is slow (sp. 2)” means that the faster copy in one species is the slower-evolving copy in the other. “Shared branch is fast” means that the shared branch leading to the consistently faster orthologs is the faster of the two shared branches. “Shared branch is slow” means that the shared branch leading to the consistently faster orthologs is the slower shared branch.
Including all the loci with nonsignificant asymmetry (data set, 2:2 loci from set 2; results, supplemental Table 2A at http://www.genetics.org/supplemental/) or applying even more stringent cutoffs (data set, 2:2 loci from set 2 with R′ > 1.25 and an absolute KA difference > 0.1; results, supplemental Table 2B at http://www.genetics.org/supplemental/) does not change the observed trends or their significance. Reducing the data set only to loci where the synteny and phylogeny agree perfectly causes no qualitative difference in our results (data set, 2:2 loci from the EQL data set in supplemental Table 1 at http://www.genetics.org/supplemental/; results, supplemental Table 2C at http://www.genetics.org/supplemental/), although significance decreases in comparisons where statistical power is greatly reduced. To rule out the possibility that the ohnolog rate asymmetry we observe is due to recombination or gene conversion of the “faster-evolving” ohnolog with a member of a paralogous family, we excluded all ohnologs with homology (BLASTP E-value <1e-20) to any other gene(s) in their genome. This step excluded 31–35% of the ohnolog pairs in the three genomes. Repeating the analyses without these loci makes no qualitative difference to the results (data not shown).
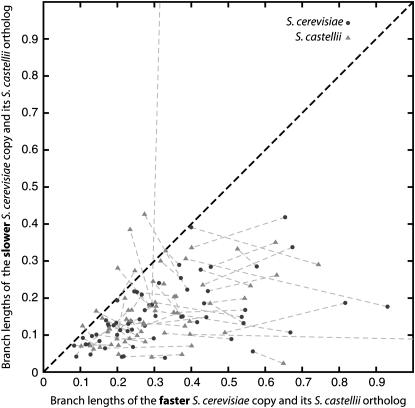
As an illustration of the consistent direction of asymmetry across species, Figure 4 compares the terminal (species-specific) branch lengths in S. cerevisiae and S. castellii at each of the 62 loci that are significantly asymmetric in both species in the 2:2 category (the same data set as in Table 1A). Each circle shows the branch lengths for the faster-evolving and the slower-evolving ohnologs in S. cerevisiae plotted against each other, and each of the triangles plots the KA-values for the orthologs in S. castellii of each of those ohnologs, with dashed lines joining the corresponding pairs. The seven triangles above the diagonal line in Figure 4 represent the small fraction of loci (11%) where the faster-evolving ohnolog is different in each species, while the remainder shows consistent asymmetry.
Figure 4.—
Consistent rate asymmetry across species. A plot of branch lengths (KA units) for the 62 significantly asymmetric 2:2 loci from the Scer–Scas species comparison in set 3 (the same data set as in Table 1) is shown. Each circle shows the branch lengths for faster (x-axis) and slower (y-axis) gene copies in S. cerevisiae, corresponding to branches A1 and B1, respectively, in Figure 1B. Triangles connected by dashed lines to each circle show the corresponding data for S. castellii, but the value of branch A2 (i.e., the S. castellii branch orthologous to the faster copy in S. cerevisiae) is always plotted on the x-axis, and the value of branch B2 (i.e., the S. castellii branch orthologous to the slower copy in S. cerevisiae) is always plotted on the y-axis, regardless of which of A2 and B2 is actually the longer branch in S. castellii. Hence any triangle lying above the diagonal is a locus where a different ohnolog is faster evolving in each species. A small number of points with branch lengths >1 are not shown.
Asymmetry is established early after duplication:
The consistent asymmetry across post-WGD species, even after shared evolutionary history has been excluded, suggests that an asymmetric evolutionary “trajectory” may have been established before speciation. To explore this further we inspected the shared branches at 2:2 loci (e.g., branches AS and BS in Figure 1B) to see if there is a significant correlation between asymmetry in amino acid distances on the shared and postspeciation branches. Examining the shared branches leading to consistently faster-evolving ohnologs [Fast (sp. 1) is Fast (sp. 2) row in Table 1A], we find a significant trend for that branch to be the faster of the two shared branches (P < 0.001; Table 1B). The bias of fast over slow is consistent and always >4:1, suggesting that asymmetric evolution began before speciation. We have previously shown that the post-WGD species considered here diverged soon after polyploidization (Scannell et al. 2006), so the asymmetric sequence divergence must itself have begun soon after the genome duplication. The same study also reported a significant overrepresentation of convergent gene losses after the speciation of the three post-WGD genomes, which is also consistent with an early established evolutionary trajectory.
The faster-evolving ohnolog is never essential and is often less well characterized:
One of the questions we address is whether the slow-evolving ohnologs tend to maintain a more essential ancestral function while the faster-evolving ohnologs have been freed to “experiment” and potentially evolve a new function, as predicted by the neofunctionalization model. We never find a fast copy of a duplicate that is essential. For example, for the 166 loci in Figure 3A, we find no fast-evolving ohnologs that are essential whereas we do see 7 loci where the slower-evolving ohnologs are essential. Even with this small number of loci the trend is significant (P = 0.015 for Fisher's exact test). For the other 2:2 comparison featuring S. cerevisiae (which is the only species for which genomewide knockout data is available) the result is consistent and also significant (Scer–Cgla, 0/136 vs. 8/128; P = 0.007). In the 2:1 and 1:2 categories we see no examples of essential genes being faster evolving but we also see only one and two examples of slower-evolving ohnologs being essential, so the results are not statistically significant.
For the most asymmetrically evolving loci (R′ > 2; 45 loci; Figure 3A), if one of the ohnologs is uncharacterized it is almost always the faster-evolving ohnolog (14 vs. 1; P < 0.05 by Fisher's exact test). Uncharacterized genes are those for which almost no functional information is available in the SGD database (Christie et al. 2004). Examples of ohnolog pairs consisting of one slow-evolving and well-characterized gene and one fast-evolving and uncharacterized gene include the known and putative pseudouridine synthases PUS1 and PUS2 (R′ = 8.5), the sorting nexin gene VPS5 and its uncharacterized ohnolog YKR078W (R′ = 5.3), and the well-characterized kinase NPR1 and its poorly understood ohnolog PRR2 (R′ = 5.7). For each of these pairs the rate asymmetry is highly significant (P < 1e-20).
Duplicates lost from one species are faster evolving in other species:
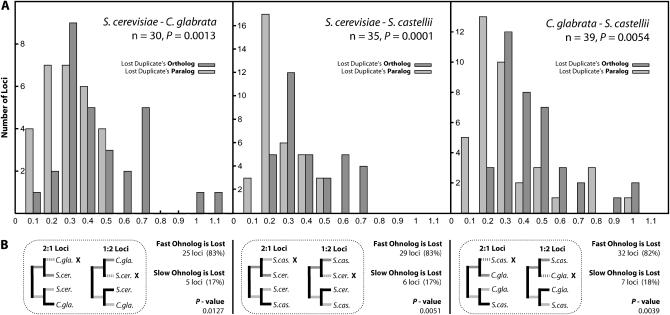
We noted an apparent trend that, at loci where one species retained only one ohnolog but other specie(s) retained pairs, the lost gene tended to be orthologous to the faster-evolving member of the pair. An example is the locus shown in Figure 2, where C. glabrata has lost its ortholog of the fast-evolving gene REG2 and retained its ortholog of the slower gene REG1. To investigate this further we tested for an association between increased rate of substitution in some ohnolog copies and loss of the orthologous copy in other species. We used the 2:1 and 1:2 categories (examining only loci that have significantly asymmetric rates of amino acid divergence; set 3; see materials and methods) to ask if the copy of the ohnolog that is lost in one species is faster evolving in a second species (e.g., in Figure 1C, where gene copy A has been lost in C. glabrata, we would test whether the branch A1 is longer than the sum of the B1 and BS branches). If so, the orthologs of lost duplicates should be faster evolving than the paralogs of lost duplicates. Comparing the rate of amino acid divergence on the branches leading to both ohnologs in the two-copy species, we find this to be the case (Figure 5A). The branches leading to the orthologs of lost genes are significantly longer than the branches leading to the paralogs of the same lost genes (P < 0.01 by paired Wilcoxon's signed rank tests on distributions for each comparison in Figure 5A). There is no qualitative difference to our results when we also include loci with nonsignificant rate asymmetry (set 2; supplemental Figure 1 at http://www.genetics.org/supplemental/).
Figure 5.—
Association across species of fast-evolving and lost ohnologs. (A) Distributions of rates of amino acid divergence (KA) on branches leading to orthologs of a lost duplicate (dark shading) are significantly greater than that on branches leading to the paralog of the lost duplicate/ortholog of the retained duplicate (light shading), in the three pairwise comparisons Scer–Cgla, Scer–Scas, and Cgla–Scas, over loci in 2:1 and 1:2 categories from set 3. P-values are from paired Wilcoxon's signed rank tests on the underlying data, with data binned for display only. We tested the hypothesis that the “lost duplicate's ortholog” KA distribution is greater than the “lost duplicate's paralog” KA distribution. (B) The 2:1 and 1:2 locus classes and the losses at them are illustrated under the distributions for each species comparison in A. The estimated KA on branches with dark shading (leading to ortholog of loss) or light shading (leading to paralog of loss) becomes part of the corresponding darkly shaded or lightly shaded distribution. The numbers and percentages to the right of each box show the trend for the fast-evolving copy in the two-copy species to be lost from the single-copy species. “Fast ohnolog is lost” means the faster copy of the gene has been lost in the other species. “Slow ohnolog is lost” means the opposite. P-values are from Fisher's exact two-tail tests against neutral expectation.
We considered that the rate asymmetry we observed between orthologs and paralogs of lost duplicates might be due to ongoing pseudogenization of the ohnolog that has been lost from other species. Using genomic data from S. bayanus (a sensu stricto species that diverged from the S. cerevisiae lineage much more recently than C. glabrata or S. castellii) we used YGOB to identify the syntenic ortholog in S. bayanus of the S. cerevisiae gene that is the ortholog of the duplicate lost from the other species in the Scer–Cgla 2:1 and Scer–Scas 2:1 categories. Using the PAML program yn00 (Yang 1997) we estimated the synonymous and nonsynonymous substitution rates between the S. cerevisiae and S. bayanus orthologs of the lost duplicate. In all cases we find a high level of constraint (ω < 0.3), indicating that the surviving orthologs of lost duplicates are still subject to purifying selection even though they have evolved quickly.
Another way to express this trend is from the perspective of the two-copy species. The numbers and percentages in Figure 5B count the number of cases where the lost copy of a gene in the one-copy species is the ortholog (“fast ohnolog is lost”) or the paralog (“slow ohnolog is lost”) of the faster-evolving ohnolog in the two-copy species. The lost copy is more often the ortholog rather than the paralog of the faster copy in any other species that retains both copies, by a factor of over fourfold (P < 0.05; Figure 5B).
DISCUSSION
As a mechanism of duplicate preservation neofunctionalization makes a number of predictions that our results endorse. Ohno (1970) predicted that when one copy of a duplicate gene pair acquires a selectively advantageous novel function and is maintained in a genome, the duplicates will evolve asymmetrically, with the neofunctionalized copy experiencing accelerated sequence evolution compared to its paralog. Our observation of widespread asymmetric sequence divergence in yeast ohnologs (Figure 3A) is consistent with extensive neofunctionalization and agrees with previous work (Conant and Wagner 2003; Kellis et al. 2004; Fares et al. 2006), confirming that most duplicated loci have evolved at least somewhat asymmetrically and many have evolved extremely asymmetrically.
It should be noted that asymmetric rates of ohnolog evolution can also be caused by asymmetric subfunctionalization arising due to an uneven partitioning of functions between the duplicates and hence differing levels of functional constraint (He and Zhang 2005). However, the large population sizes of yeasts mean that subfunctionalization is not expected to be an important mechanism of duplicate retention (Lynch and Force 2000a). Although rate asymmetry itself is not conclusive evidence of neofunctionalization, the overall patterns of asymmetric evolution that we observe in yeasts are highly suggestive of neofunctionalization.
The significant asymmetry observed in amino acid substitution rates for 56% of ohnolog pairs is strikingly higher than the 30% previously reported for duplicates in a number of species (Conant and Wagner 2003). This difference may be due to our use of interspecific outgroup genes or our use of phylogenetic filters to remove duplicates that appear to have undergone gene conversion and that would not be expected to show evidence of asymmetric evolution. It is apparent that a very large fraction of the ohnolog pairs preserved in degenerate polyploid yeast genomes have evolved significantly asymmetrically.
Our discovery that the direction of asymmetry is consistent over evolutionary time (i.e., on the branches before and after speciation; AS/BS, A1/B1, and A2/B2 in Figure 1B) is characteristic of neofunctionalization for several reasons. First, asymmetric evolution is a consequence of the action of neofunctionalization as a mechanism of duplicate preservation and so is expected to have begun shortly after polyploidization. The significant correlation of the faster shared branch and the consistently faster-evolving ohnolog (Table 1B) offers direct support that this is what has happened. We have previously shown that the post-WGD species diverged soon after polyploidization (Scannell et al. 2006), so the asymmetric sequence divergence must itself have begun soon after the genome duplication. Second, the early establishment of an asymmetric evolutionary trajectory also means that the identity of the faster-evolving ohnolog is expected to be the same across species. The observation that, even after controlling for shared evolutionary history, the faster-evolving ohnologs in one species are significantly more likely to be faster evolving in other species (∼90% of 2:2 loci; Table 1A) shows that this is indeed the case. Although asymmetric subfunctionalization can also result in evolutionary rate asymmetry (He and Zhang 2005) neither of these trends is expected under subfunctionalization because asymmetry is not a consequence of that preservation mechanism, but something that may happen later. After subfunctionalization, accelerated evolution, if it occurs, could occur in different copies of the duplicate in different species, or in only one species, rather than consistently in the same copy across species as we observe.
Our results indicate that neofunctionalization occurred soon after duplication and was widespread. Was neofunctionalization also the initial mechanism of preservation of the gene pairs? In principle, the rate asymmetries we observe could alternatively have been caused by neofunctionalization occurring at loci that had already been preserved in duplicate either by rapid subfunctionalization (He and Zhang 2005) or by dosage selection (Sugino and Innan 2006). However, duplicate preservation by neofunctionalization also predicts that while one of the ohnologs retains an ancestral function the other is relieved of selective constraint, becomes free to evolve more rapidly, and potentially acquires a new function. Our finding that faster-evolving ohnologs are never essential and often uncharacterized provides support that this is what has happened in yeast ohnologs. The trend is again more suggestive of neofunctionalization than of subfunctionalization, because preservation due to subfunctionalization strongly rules out the possibility of future loss of either member of the pair; the partitioning of important ancestral subfunctions, which is integral to the initial preservation of the pair, cannot easily be reversed. Yet we see such losses, and, as predicted under neofunctionalization, they are predominantly losses of orthologs of the faster-evolving members of the ohnolog pair (by a factor of 4:1; Figure 5B). Similarly after neofunctionalization the ancestral function of the slow-evolving gene is more likely to have been characterized than any novel function the fast-evolving ohnolog may have acquired.
Although we can rule out the partitioning of discrete ancestral functions (i.e., classical subfunctionalization) as a common mechanism of yeast ohnolog preservation, we cannot rule out preservation for dosage reasons followed by later neofunctionalization. In particular, it is possible that preservation for dosage can be followed later by shifts in the expression levels of an ohnolog pair, resulting in a highly expressed gene copy that retains the ancestral function and a lowly expressed copy that may become neofunctionalized. This idea is explored elsewhere (D. R. Scannell and K. H. Wolfe, unpublished data).
In conclusion, the consistent patterns of rate asymmetry and loss that we observe in yeast ohnologs demonstrate that neofunctionalization was widespread soon after WGD. The large proportion of neofunctionalized loci and the later loss of some faster-evolving ohnologs further suggest that neofunctionalization may also have been the method of duplicate preservation at many loci.
Acknowledgments
We thank Meg Woolfit for critical comments on the manuscript and Gavin Conant, David Lynn, Devin Scannell, and Brian Cusack for helpful discussion. This study was supported by Science Foundation Ireland.
References
- Benjamini, Y., and Y. Hockberg, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, K. P., and G. Blanc, 2006. Computational analyses of ancient polyploidy. Curr. Bioinform. 1: 131–146. [Google Scholar]
- Byrne, K. P., and K. H. Wolfe, 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15: 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes, J. K., G. P. Morris and W. H. Li, 2006. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol. Biol. Evol. 23: 1136–1143. [DOI] [PubMed] [Google Scholar]
- Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson et al., 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, K. R., S. Weng, R. Balakrishnan, M. C. Costanzo, K. Dolinski et al., 2004. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32: D311–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al., 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Cliften, P. F., R. S. Fulton, R. K. Wilson and M. Johnston, 2006. After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics 172: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant, G. C., and A. Wagner, 2003. Asymmetric sequence divergence of duplicate genes. Genome Res. 13: 2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates et al., 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304: 304–307. [DOI] [PubMed] [Google Scholar]
- Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola et al., 2004. Genome evolution in yeasts. Nature 430: 35–44. [DOI] [PubMed] [Google Scholar]
- Fares, M. A., K. P. Byrne and K. H. Wolfe, 2006. Rate asymmetry after genome duplication causes substantial long branch attraction artifacts in the phylogeny of Saccharomyces species. Mol. Biol. Evol. 23: 245–253. [DOI] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, D. L., and K. Tatchell, 1996. The REG2 gene of Saccharomyces cerevisiae encodes a type 1 protein phosphatase-binding protein that functions with Reg1p and the Snf1 protein kinase to regulate growth. Mol. Cell. Biol. 16: 2922–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon et al., 1996. Life with 6000 genes. Science 274: 546, 563–567. [DOI] [PubMed]
- Guldener, U., M. Munsterkotter, G. Kastenmuller, N. Strack, J. van Helden et al., 2005. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 33: D364–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., and J. Zhang, 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H., K. Tatchell, S. Liu and C. A. Michels, 2000. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol. Gen. Genet. 263: 411–422. [DOI] [PubMed] [Google Scholar]
- Kaniak, A., Z. Xue, D. Macool, J. H. Kim and M. Johnston, 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., B. W. Birren and E. S. Lander, 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Krakauer, D. C., and M. A. Nowak, 1999. Evolutionary preservation of redundant duplicated genes. Semin. Cell Dev. Biol. 10: 555–559. [DOI] [PubMed] [Google Scholar]
- Li, W. H., 1997. Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Lynch, M., and A. Force, 2000. a The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and A. G. Force, 2000. b The origin of interspecies genomic incompatibility via gene duplication. Am. Nat. 156: 590–605. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and V. Katju, 2004. The altered evolutionary trajectories of gene duplicates. Trends Genet. 20: 544–549. [DOI] [PubMed] [Google Scholar]
- Maere, S., S. De Bodt, J. Raes, T. Casneuf, M. Van Montagu et al., 2005. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102: 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. George Allen & Unwin, London.
- Papp, B., C. Pal and L. D. Hurst, 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., B. A. Chapman, J. C. Kissinger, J. E. Bowers, F. A. Feltus et al., 2006. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 22: 597–602. [DOI] [PubMed] [Google Scholar]
- Phillips, M. J., F. Delsuc and D. Penny, 2004. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 21: 1455–1458. [DOI] [PubMed] [Google Scholar]
- Scannell, D. R., K. P. Byrne, J. L. Gordon, S. Wong and K. H. Wolfe, 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440: 341–345. [DOI] [PubMed] [Google Scholar]
- Sugino, R. P., and H. Innan, 2006. Selection for more of the same product as a force to enhance concerted evolution of duplicated genes. Trends Genet. 22: 642–644. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Ter Linde, J. J., and H. Y. Steensma, 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19: 825–840. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., J. S. Taylor, I. Braasch and A. Meyer, 2001. The ghost of selection past: rates of evolution and functional divergence of anciently duplicated genes. J. Mol. Evol. 53: 436–446. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., 2005. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171: 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K., 2000. Robustness—it's not where you think it is. Nat. Genet. 25: 3–4. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713.9192896 [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang, L., T. J. Vision and B. S. Gaut, 2002. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 19: 1464–1473. [DOI] [PubMed] [Google Scholar]