Abstract
Regulation of gene transcription is a key feature of developmental, homeostatic, and oncogenic processes. The reverse recruitment model of transcriptional control postulates that eukaryotic genes become active by moving to contact transcription factories at nuclear substructures; our previous work showed that at least some of these factories are tethered to nuclear pores. We demonstrate here that the nuclear periphery is the site of key events in the regulation of glucose-repressed genes, which together compose one-sixth of the Saccharomyces cerevisiae genome. We also show that the canonical glucose-repressed gene SUC2 associates tightly with the nuclear periphery when transcriptionally active but is highly mobile when repressed. Strikingly, SUC2 is both derepressed and confined to the nuclear rim in mutant cells where the Mig1 repressor is nuclear but not perinuclear. Upon derepression all three subunits (α, β, and γ) of the positively acting Snf1 kinase complex localize to the nuclear periphery, resulting in phosphorylation of Mig1 and its export to the cytoplasm. Reverse recruitment therefore appears to explain a fundamental pathway of eukaryotic gene regulation.
THE nonrandom distribution of chromatin was first noted >100 years ago (Rabl 1885). The advent of molecular biology has provided a plausible explanation for this interesting observation—in eukaryotes the nuclear position of a gene may determine its transcriptional status. The nucleus is now known to be divided into distinct chromosomal and proteinaceous subcompartments, including interchromatin granule clusters (IGCs), promyelocytic leukemia (PML) bodies, Cajal bodies, SC35 complexes, and the classically described nucleoli and nuclear pore complexes (NPCs) (Pederson 2002; Lamond and Sleeman 2003). Each of these ultrastructural features has been implicated in various steps in gene expression, including mRNA export, splicing, and transcription initiation (Smith et al. 1999; von Mikecz et al. 2000; Maniatis and Reed 2002; Murphy et al. 2002; Sacco-Bubulya and Spector 2002; Granneman and Baserga 2005; Menon et al. 2005; Cabal et al. 2006; Schmid et al. 2006; Taddei et al. 2006). This has led to the idea that at least a subset of RNA polymerase II (Pol II) complexes is organized into gene expression machines (Maniatis and Reed 2002; Santangelo 2006) or transcription factories (Cook 2001). However, definitive experimental verification of a molecular model for eukaryotic gene regulation that is predicated upon the existence of nuclear ultrastructure has long been elusive.
Reverse recruitment, a recently proposed molecular model for eukaryotic gene regulation, postulates that genes become active by moving to contact transcription factories that are localized to nuclear substructures and that at least some of these factories are tethered to NPCs (Menon et al. 2005). This model was first used to explain the Rap1/Gcr1 activation mechanism, which accounts for ≥75% of mRNA generation in rapidly growing cells of the budding yeast Saccharomyces cerevisiae (Santangelo 2006; Barbara et al. 2007). Transcriptomics and other analyses have also implicated the perinuclear regulator Gcr1 in glucose repression (Turkel et al. 2003; Sasaki and Uemura 2005), a result that the reverse recruitment paradigm can accommodate in a straightforward and parsimonious fashion (Santangelo 2006).
Our previous work suggested that Rap1/Gcr1 transcriptional activation requires the integrity of an NPC-anchored assemblage that provides ready access to active Pol II complexes (Menon et al. 2005). The existence of a functional relationship between Pol II and NPCs is also indicated by nucleoporin activation, which is easily detected in one-hybrid assays. Since promoters lacking binding sites for transcriptional activators fail to drive transcription in vivo, and the default state of chromatin is silent, the capacity of nucleoporins to function as activators is highly significant. Interestingly, we have found that nucleoporin activation is glucose repressed. We therefore investigated the potential applicability of reverse recruitment to the glucose repression regulatory pathway. This pathway is a particularly robust system with which to test models for gene regulation, since the roles of the key regulatory proteins responsible for genomewide repression and derepression are very well established (for recent review see Santangelo 2006). As in mammalian cells, signal transduction through the Ras/cAMP-dependent protein kinase A (PKA) pathway initiates a transcriptomewide glucose response (Wang et al. 2004). The negative regulators Hxk2, Mig1, and Ssn6 respond to this signal by collaborating to block transcription of glucose-repressed genes (Carlson 1999). Derepression in the absence of glucose requires phosphorylation of the zinc finger-containing DNA-bound repressor Mig1 by the Snf1 kinase complex (Treitel et al. 1998). We report here that the reverse recruitment model explains key features of the Hxk2/Mig1/Ssn6/Snf1 glucose repression pathway. All three subunits of the Snf1 kinase are perinuclear when they are needed to counteract glucose repression by Hxk2/Mig1. Importantly, the canonical target gene SUC2 is highly mobile and randomly positioned in the yeast nucleus when repressed, but associates tightly with NPCs when derepressed.
MATERIALS AND METHODS
Growth and assays:
Cells were grown in synthetic complete medium, to which 2% glucose (repressing conditions) or 3% pyruvate (derepressing conditions) was added as carbon source, unless otherwise indicated. Standard assays were done to measure β-galactosidase activity (Zeng et al. 1997).
Cell fractionation and QFPD assay:
Cytosolic, nuclear (nucleoplasmic/perinuclear), and perinuclear fractions were isolated as previously described (Kipper et al. 2002). α-Snf1 (Santa Cruz Biotechnology) and α-Pom152 (a generous gift of Michael Rout) were used in Western analysis. Strains from the Yeast GFP Clone collection (Invitrogen Life Technologies, San Diego) were used in quantitative fluorescent protein detection (QFPD) experiments. QFPD assays involved loading a portion of each of the aforementioned fractions (0, 2, 4, 8, and 16 μg for Htb2-GFP and Nog2-GFP; 0, 20, 40, 80, and 160 μg for all others) into 96-well plates for analysis with a Typhoon Phosphorimager (GE Healthcare). GFP was excited by using the 488-nm laser and the resulting fluorescence was acquired with the 526 short-pass emission filter at high sensitivity with detection at +3 mm above the platen surface at 200-μm resolution. For quantitative analysis, densitometric values were obtained by using ImageQuant (GE Healthcare) and units of GFP per milligram of protein were determined after normalization.
Confocal laser scanning microscopy:
For in vivo time-lapse microscopy, a Zeiss LSM510 META confocal microscope with a 100× αPlan-Fluar 1.45 NA oil objective lens was used to capture 10 series of nuclei from cells grown on selective SC media plates containing either 2% glucose or 3% pyruvate. Wild-type (WT) cells (isogenic to BY263, with 256 repeats of the Lac operator integrated at position −1500 to the SUC2 gene) contained plasmid-borne LacI-GFP (probe) and Nsp1-yellow fluorescent protein (YFP) (peripheral marker), HIS3 and URA3 marked, respectively. GFP and YFP were excited with the 488- and 514-nm lasers and detected with 505–530 BP and 530 LP filters, respectively. Imaging was done using an αPlan-Fluar 100×/1.45 NA objective with a depth of focus of 1 μm; resolution was 0.04 μm/pixel. Time lapse was performed over 4 min with an image taken every 60 sec starting at time zero. Each P-value associated with the localization of SUC2 within the nucleus was calculated by using an unpaired Student's t-test.
Chromatin immunoprecipitation:
Chromatin extracts were prepared from TAP-tagged and HA-tagged strains (Open Biosystems) as described (Boukaba et al. 2004). Immunoprecipitation was done with 5 μg of α-TAP (Open Biosystems) or α-HA (12CA5; Roche Molecular Biochemicals, Indianapolis) antibody and protein-A Sepharose beads, using the method detailed in Boukaba et al. (2004). The final DNA pellet was resuspended in 30 μl TE; 1, 2, and 3 μl were used for PCR amplification of target regions. PCR detection of the SUC2 and ACT1 promoters was done with primers specific to the −500- to −850-bp region. Twenty percent of PCR products were resolved on 2% Nusieve agarose gels and imaged with a Chemidoc XRS (Bio-Rad, Hercules, CA).
RESULTS
Snf1-dependent derepression of nucleoporin activators:
We previously used a standard lexA one-hybrid assay to show that components of the Nup84 subcomplex (Siniossoglou et al. 2000) function as potent transcriptional activators while remaining properly localized to the nuclear rim (Menon et al. 2005). Interestingly, this nucleoporin activation, which spans a range of ∼3 logs of activity, is repressed by glucose; six different NPC subunits are weaker activators in the presence of glucose (Table 1). Western blots confirmed that glucose-repressed reporter gene activity was not due to a carbon source-dependent change in the cellular levels of nucleoporin-lexA chimeras (supplemental Figure 1 at http://www.genetics.org/supplemental/ and data not shown). A Ste12-lexA chimera was tested as an additional control. Ste12 is a conventional activator known to be governed by MAP kinase in the regulatory pathways that control mating and pseudohyphal growth (Pi et al. 1997); both its activity and its protein level were found to be unaffected by the presence or absence of glucose (Table 1 and data not shown).
TABLE 1.
Nucleoporin activation is repressed by glucose
| lexA fusiona | R (glucose) | D (no glucose) | D/R ratiob |
|---|---|---|---|
| NUP84 | 2303c | 5185c | 2.3 |
| NUP145C | 799 | 3403 | 4.3 |
| NUP120 | 191 | 766 | 4.0 |
| NUP145N | 65 | 521 | 8.0 |
| SEC13 | 17 | 87 | 5.1 |
| NUP133 | 7 | 57 | 8.1 |
| STE12 | 124 | 133 | 1.1 |
| lexA alone | <0.1 | <0.1 | — |
C-terminal fusion to the lexA DNA-binding domain. The classically described STE12 activator and lexA DNA-binding domain alone are shown as controls.
Ratio of derepressed to repressed reporter gene expression.
lexA-driven β-galactosidase activity. Four independent determinations of β-galactosidase activity (units per milligram of total protein) were averaged; standard error of the mean for each value was <10%.
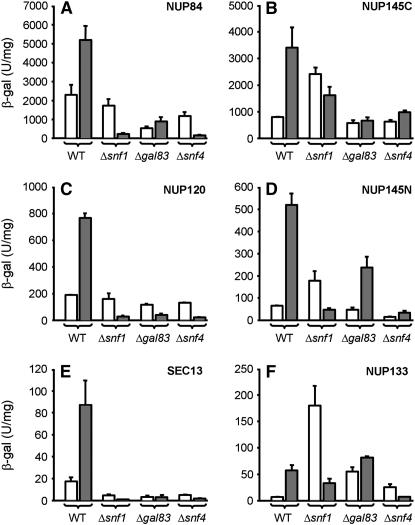
The Snf1 kinase complex is required for derepression of glucose-repressed genes (Treitel et al. 1998). This complex, like its mammalian counterpart (the AMP-activated protein kinase, AMPK) (Hardie et al. 1998), is heterotrimeric and phosphorylates serines and threonines in targeted regulators in the yeast nucleus such as the glucose-responsive repressor Mig1 (Treitel et al. 1998). We therefore investigated whether nucleoporin activation in the absence of glucose requires Snf1 kinase subunits. SNF1 encodes the catalytic α-subunit, GAL83 encodes the most abundant β-subunit, and SNF4 encodes the γ-subunit (Vincent et al. 2001). We tested nucleoporin-lexA chimeras for Snf1 dependence by introducing them into Δsnf1, Δgal83, or Δsnf4 reporter strains; we then grew the cells in either the presence or the absence of glucose and assayed for β-galactosidase reporter gene activity. Western blots confirmed that removal of Snf1, Gal83, or Snf4 had no effect on expression of the nucleoporin-lexA polypeptides (supplemental Figure 1 at http://www.genetics.org/supplemental/ and data not shown). As anticipated, derepression of transcription mediated by each of the six glucose-regulated nucleoporins was defective in the absence of the α-, the β-, or the γ-subunit of the Snf1 kinase (Figure 1, A–F, shaded bars). Importantly, for two nucleoporins (Nup145C and Nup145N) both derepression and repression were defective in the absence of the catalytic (Snf1) α-subunit (Figure 1, B and D); for a third (Nup133), repression was defective in the absence of either the α- or the β-subunit (Figure 1F). The latter unexpected results are a further suggestion that glucose derepression and repression may functionally overlap to a greater degree than previously appreciated (Santangelo 2006).
Figure 1.—
Derepression of nucleoporin-mediated transcription requires the Snf1 kinase complex. Wild type (WT), Δsnf1, Δgal83, and Δsnf4 strains containing a chromosomally integrated lexA-driven reporter gene were transformed with nucleoporin-lexA fusion genes as described previously and assayed for β-galactosidase activity after growth in either the presence (open bars, repression) or the absence (shaded bars, derepression) of glucose. Each of the nucleoporin-lexA fusion genes tested in this assay was expressed at normal levels, i.e., was driven by its native promoter. LexA fusion genes are (A) NUP84, (B) NUP145C, (C) NUP120, (D) NUP145N, (E) SEC13, and (F) NUP133. Error bars represent the standard deviation of four independent determinations. Reporter expression driven by control proteins, which included Ste12-lexA and Gcn4-lexA chimeras, was similar in the presence or absence of Snf1 kinase subunits.
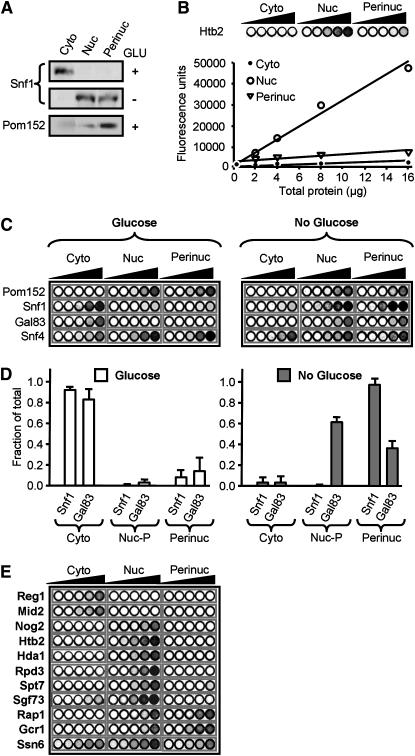
Snf1-dependent derepression of nucleoporin activation suggested a functional and perhaps even a physical connection between the kinase complex and the nuclear rim. To test for a physical connection, we isolated cytoplasmic, nuclear, and perinuclear fractions as described previously (Kipper et al. 2002; Menon et al. 2005) and assayed for Snf1 in these different compartments; Pom152, an NPC-specific integral nuclear membrane protein, was used as a perinuclear control (Menon et al. 2005). Figure 2A shows that in the absence of glucose Snf1 is nuclear (as previously reported; Schmidt and McCartney 2000; Vincent et al. 2001) and exhibits significant enrichment in perinuclear fractions.
Figure 2.—
The active Snf1 kinase complex cofractionates with perinuclear components. Cyto, cytoplasmic fraction; Nuc, combined nucleoplasmic/perinuclear fraction; Perinuc, perinuclear fraction; Glu or Glucose, extracts from glucose-grown cells. (A) (Top and middle) Western blot of Snf1; (bottom) Western blot of Pom152. (B) Quantitative fluorescent protein detection (QFPD) assay. Increasing amounts of total protein from indicated fractions containing H2B-GFP were loaded into microtiter wells (circles, left to right), and fluorescence was measured as described in materials and methods; the graph shows the corresponding densitometric analysis. (C) QFPD analysis of Snf1-GFP, Snf4-GFP, and Gal83-GFP as in B above. (D) Densitometric analysis of the data shown in C. The fraction of total Snf1-GFP and Gal83-GFP fluorescence present in cytoplasmic (Cyto), nucleoplasmic (Nuc-P, total nuclear fluorescence minus perinuclear fluorescence), and perinuclear (Perinuc) fractions is represented. Open bars, glucose; shaded bars, no glucose. Error bars represent the standard error of the mean. (E) QFPD assay of nuclear proteins. Cytoplasmic Reg1 and Mid2, nucleolar Nog2, and perinuclear Rap1 and Gcr1 are shown as controls.
To test whether or not the remaining Snf1 kinase subunits, as well as other transcription factors, localize to the perinuclear compartment, we developed a sensitive new laser-based method for QFPD (Figure 2, B–E). This method combines the above biochemical fractionation technique with fluorescence-based quantitation of GFP chimeras. The availability of a GFP collection of 4156 yeast strains, each of which expresses a different full-length, chromosomally tagged GFP chimera (Huh et al. 2003), allows rapid and comprehensive analysis of the cytoplasmic, nuclear, and perinuclear distribution of virtually any factor in yeast cells.
As a proof of concept for QFPD we again isolated biochemical fractions corresponding to the cytoplasmic, nuclear, and perinuclear compartments, this time by using a strain that contained GFP-tagged histone 2B (H2B, encoded by HTB2). Increasing amounts of protein from each fraction were loaded into a 96-well plate, which was scanned with the 488-nm laser of a Typhoon phosphorimager. There was a linear relationship between the amount of total protein tested and the intensity of H2B-GFP fluorescent emission, allowing quantitation over a ≥10-fold range (Figure 2B). Since DNAse treatment was used to release chromatin-bound factors prior to isolation of perinuclear fractions (Kipper et al. 2002; Menon et al. 2005), it was not surprising to find that H2B-GFP is a nuclear factor that exhibits little if any perinuclear association (Figure 2B). Chromatin factors thus serve as an excellent negative control in determining how much of a given yeast regulator is present in the perinuclear compartment. Additional examples include the histone deacetylases Hda1 and Rpd3, as well as the SAGA complex components Spt7 and Sgf73, none of which are detectable in perinuclear fractions (Figure 2E). As a further test of the QFPD method, we analyzed strains containing GFP-tagged Rap1, Gcr1 (Figure 2E), and Pom152 (Figure 2C); the QFPD results for these factors matched previously published Western blot data (Menon et al. 2005).
We used QFPD to test localization of all three subunits of the Snf1 kinase complex in cells grown in either the presence or the absence of glucose. Whereas Snf4-GFP is present in both the cytoplasm and the nucleus irrespective of the carbon source (Figure 2C), both Snf1-GFP and Gal83-GFP are cytoplasmic in the presence of glucose and nuclear in its absence (Figure 2, C and D). These data agree with previous analyses of the subcellular localization of Snf1 kinase subunits with fluorescence microscopy (Schmidt and McCartney 2000; Vincent et al. 2001). Importantly, glucose-grown cells contain little if any detectable perinuclear Snf1 or Gal83; in contrast, in the absence of glucose cells contain a substantial fraction of all three subunits in the perinuclear compartment (Figure 2, C and D). This served as a further validation of the QFPD method, since the result for Snf1-GFP (Figure 2, C and D) was identical to that obtained by using a native α-Snf1 antibody in Western blot analysis (Figure 2A). Thus derepression of nucleoporin-stimulated transcription requires the entire Snf1 kinase complex, and both the kinase and its regulatory β-subunit cofractionate with NPCs only when they are needed to counteract glucose repression in the yeast nucleus.
Glucose derepression involves tight association between SUC2 and the nuclear periphery:
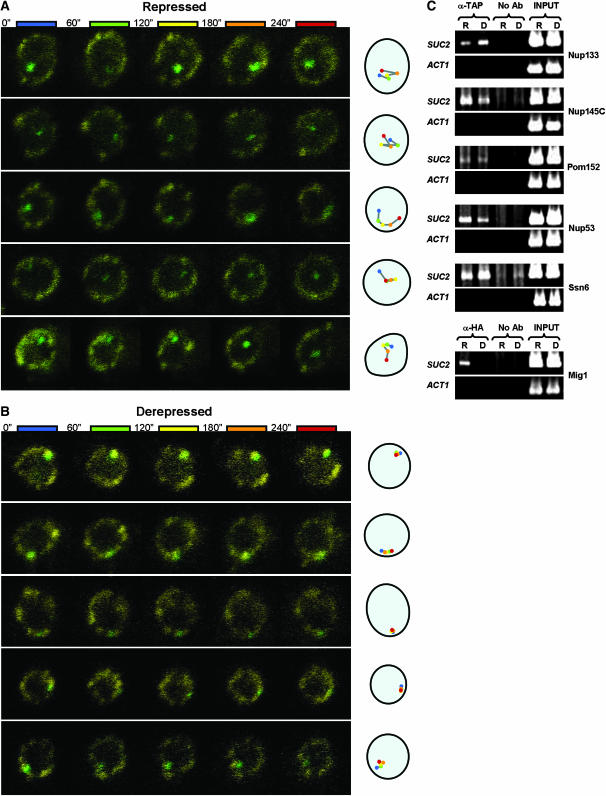
Recent work has clearly demonstrated that transcriptionally active target genes in S. cerevisiae (e.g., those regulated by Rap1/Gcr1, Gal4, and Ino2) physically associate with perinuclear factors (Brickner and Walter 2004; Casolari et al. 2004, 2005; Menon et al. 2005). Given the data presented above, we suspected that SUC2, the canonical glucose-repressed locus on chromosome IX, might also be associated with the nuclear periphery when active. We tested this by inserting an array of lac operators upstream of SUC2 in cells that expressed a LacI repressor fused to green fluorescent protein (LacI-GFP); this allowed monitoring of the in vivo subnuclear position of the tagged locus within a confocal optical slice. Coexpression of a YFP-tagged nucleoporin (Nsp1-YFP) allowed simultaneous visualization of the nuclear rim.
In glucose-grown cells (under repressing conditions), the location of SUC2 in the nucleus appeared to be random (Figure 3A). It was present in the perinuclear compartment (taken as the outer third of the nuclear area; see Taddei et al. 2006) in 45% of cells (n = 153). In the absence of glucose (under derepressing conditions), when invertase activity is ∼100-fold higher (Table 2), SUC2 localization to the perinuclear compartment increased to 74% (Figure 3B; n = 143; P < 0.0001). Time lapse fluorescence microscopy revealed a striking carbon source-dependent difference in the intranuclear motility of SUC2. The position of SUC2 was dynamic in glucose-grown cells (Figure 3A; supplemental movies S1–S3 at http://www.genetics.org/supplemental/). However, in the absence of glucose, movement of SUC2 was restricted to the perinuclear compartment, including occasional sliding along the nuclear periphery (Figure 3B; supplemental movies S1–S3). Although the underlying explanation for this lateral perinuclear sliding remains to be determined, similar behavior has also recently been reported for other transcriptionally active genes (Cabal et al. 2006; Taddei et al. 2006).
Figure 3.—
SUC2 exhibits carbon-source-dependent motility and associates with NPCs. Time-lapse analysis (4 min total) of the location of GFP-tagged SUC2 in either (A) repressed or (B) derepressed cells is shown; the result for each of five different nuclei in either repressing or derepressing conditions is presented horizontally in temporal order from left to right. The cartoon on the right depicts the location of the gene within the nucleus at the time indicated, in seconds: 0 (blue), 60 (green), 120 (yellow), 180 (orange), and 240 (red). A YFP fusion to the essential NPC component Nsp1 marks the nuclear periphery. (C) ChIP analysis of association between SUC2 and factors that represent different strata of the nuclear periphery. TAP-tagged Nup53 (NPC subunit), Nup133 (NPC subunit), Nup145 (NPC subunit), Pom152 (NPC-specific integral nuclear membrane protein), or Ssn6 (perinuclear Mig1 corepressor; see Figure 2E), or HA-tagged Mig1, were immunoprecipitated from cells grown on either the presence (R, repressed) or absence (D, derepressed) of glucose. The SUC2 promoter and ACT1 (negative control) promoter were amplified from both immunoprecipitated material and whole-cell extracts (INPUT, positive control). No Ab, no antibody (negative control).
TABLE 2.
Increased SUC2 expression correlates with increased localization of the gene to the nuclear periphery
|
SUC2 expressiona
|
% cells with peripheral SUC2 ORFb
|
|||
|---|---|---|---|---|
| Genotype | R (glucosec) | D (no glucosec) | R (glucosec) | D (no glucosec) |
| WT | 2.6 ± 0.2 | 267.2 ± 31.3 | 45 | 74 |
| Δmig1 | 52.8 ± 8.8 | 148.4 ± 1.8 | 60* | 69 |
| Δhxk2 | 65.4 ± 8.2 | 164.1 ± 36.6 | 60** | 72 |
SUC2 expression was measured with invertase assays as described in Neigeborn and Carlson (1984). Error denotes the standard deviation of four determinations.
Percentage of cells where the SUC2 ORF is localized to the outer third of the nucleus. P-values, calculated on the basis of a two-tailed Student's t-test, represent a significant difference in the localization of the ORF in glucose-grown WT cells relative to glucose-grown Δmig1 or Δhxk2 cells. *Significant (P < 0.01); **highly significant (P < 0.0005). At least 115 cells were measured for each condition.
See materials and methods for growth conditions.
Chromatin immunoprecipitation (ChIP) data (Figure 3C), which were verified across a three-fold range of PCR template, show that the SUC2 promoter associates with at least four different components of the NPC (Nup53, Nup133, Nup145C, and Pom152; Figure 3C) in both the presence and the absence of glucose; removal of some of these same factors impairs both repression and derepression of SUC2 (see below). SUC2 is also constitutively associated with the corepressor Ssn6 (Figure 3C), which is substantially enriched in perinuclear fractions in both the presence (Figure 2E) and the absence of glucose (data not shown). In contrast, SUC2 associates with Mig1 only in the presence of glucose; this was expected because Mig1 is exported to the cytoplasm in the absence of glucose (De Vit et al. 1997).
Mig1 repression occurs in the perinuclear compartment:
Many glucose-regulated genes are transcriptionally repressed by the DNA-bound factor Mig1 (Lutfiyya et al. 1998). In glucose-grown Δmig1 cells, invertase activity is ∼20-fold higher than in wild type (Vallier and Carlson 1994). We found that this increase in SUC2 expression corresponds with a significant increase in the peripheral localization of the ORF; the gene is localized to the nuclear periphery in 60% of Δmig1 cells (n = 146) vs. 45% of glucose-grown wild-type cells (n = 152, P < 0.01; Table 2). In agreement with previous reports (De Vit et al. 1997; DeVit and Johnston 1999), our confocal microscopy-based analysis showed Mig1-GFP to be nuclear in the presence of glucose and cytoplasmic in the absence of glucose (Figure 4A). QFPD analysis also finds that Mig1 is nuclear in glucose-grown cells (Figure 4B), i.e., when it binds upstream of SUC2 (Figure 3C) and represses transcription (Carlson 1999; Turkel et al. 2003). Interestingly, QFPD analysis further shows that the perinuclear compartment contains a substantial fraction of Mig1 (Figure 4B). This suggests that repression by Mig1 is a dynamic process that requires association with both the promoter DNA and the nuclear periphery (see discussion). In the absence of glucose (derepressing conditions), Mig1 exits the perinuclear compartment and is found predominantly in the cytoplasm (Figure 4).
Figure 4.—
Perinuclear localization is required for repression by Mig1. (A) Confocal laser scanning microscopy of Mig1-GFP in wild-type or Δhxk2 cells grown in either the presence or the absence of glucose, as indicated; coexpressed Rap1-CFP is shown as a nuclear marker. GFP and CFP signals were captured by using a Zeiss LSM 510 META confocal laser scanning microscope with a 63× Plan-Apochromat 1.4 NA oil DIC objective lens. Signals were separated with a 490-nm dichroic mirror with long pass filters adjusted to 505 and 475 nm, respectively. Pinholes were adjusted to obtain <1.8-μm optical slices. Images were acquired with the Zeiss LSM 510 software version 3.2. (B) Graphs show the fraction of total Mig1-GFP fluorescence present in cytoplasmic (Cyto), nucleoplasmic (Nuc-P, total nuclear fluorescence minus perinuclear fluorescence), and perinuclear (Perinuc) fractions, as determined by QFPD analysis. Open bars, glucose; shaded bars, no glucose. Error bars represent the standard error of the mean.
Deletion of HXK2, which encodes the predominant form of hexokinase in S. cerevisiae, has long been known to cause defects in glucose repression (Entian 1980). In Δhxk2 cells grown on glucose, invertase activity increases >70-fold (Neigeborn and Carlson 1984, 1987); this increase remains unaltered in Δhxk2 Δmig1 cells (data not shown). Thus, despite its presence in the nucleus of glucose-grown Δhxk2 cells (Figure 4A), Mig1 has lost its function as a repressor. Strikingly, QFPD analysis of Mig1-GFP demonstrated that nuclear Mig1 is depleted from the perinuclear fraction of glucose-grown Δhxk2 cells (Figure 4B), indicating that the repressor function of Mig1 requires its presence in the perinuclear compartment. Consistent with this result, SUC2 is retained at the nuclear periphery in 60% (n = 150) of glucose-grown Δhxk2 cells (Table 2), a significant increase over that in glucose-grown wild type cells (P < 0.0005). In the absence of glucose, SUC2 is retained at the periphery in 72% of Δhxk2 cells (n = 158); thus in derepressed conditions, both the perinuclear localization of the gene and expression levels (Neigeborn and Carlson 1984) are comparable in the presence and absence of Hxk2.
DISCUSSION
Our work suggests that the function of classically described positively and negatively acting regulators that respond to glucose can be explained by their movement into or out of the perinuclear compartment. This is consistent with the previously proposed reverse recruitment model for gene regulation (Menon et al. 2005). Two subunits of the derepressing Snf1 kinase complex, Snf1 itself and Gal83, are cytoplasmic under repressing conditions but are perinuclear in the absence of glucose (Figure 2, A, C, and D). Conversely, the Mig1 repressor is present in the perinuclear compartment and bound to the SUC2 promoter only in the presence of glucose (Figures 4B and 3C), and its Ssn6 corepressor is also substantially perinuclear and bound to SUC2 during repression (Figures 2E and 3C). The action of these regulators at the nuclear periphery provides an explanation for our initial observation of glucose-repressed nucleoporin activation (Figure 1). Repression of nucleoporin-activated reporter gene transcription requires the corepressor Ssn6 in the presence of glucose (data not shown) and is derepressed by the Snf1 kinase complex in the absence of glucose (Figure 1). We have also found that glucose regulation is deranged upon removal of perinuclear factors. For example, in Δnup133 cells SUC2 expression, as measured by invertase activity, is at least twofold reduced in the absence of glucose; in the presence of glucose, SUC2 expression is approximately fivefold derepressed. We observe these changes in regulation despite normal nucleocytoplasmic shuttling of Mig1 and Snf1 in Δnup133 cells (data not shown). Taken together, our data suggest that regulation of glucose-repressed yeast genes takes place at the nuclear periphery and further suggest that the Nup84 subcomplex, an essential structure within NPCs, plays a critical role.
Consistent with the perinuclear location of its regulators (Mig1, Ssn6, and Snf1), as well as the loss of glucose derepression in the absence of genes encoding perinuclear factors, SUC2 is tightly associated with the periphery when active and exhibits greatly increased motility when repressed (Figure 3, A and B; supplemental movies S1–S3 at http://www.genetics.org/supplemental/). However, despite the increased motility during repressing conditions, a physical interaction between SUC2 and NPCs can still be detected (Figure 3C). One simple explanation for this result might be that a transient random interaction is sufficient to produce a strong ChIP signal; alternatively, the gene may periodically revisit the site of regulatory action in the perinuclear compartment. We favor the latter explanation, that the repressed SUC2 gene occasionally makes physical contact with one or more perinuclear sites of activation but is unable to establish a productive interaction due to Hxk2-mediated interference by the perinuclear Mig1/Ssn6 repressor. In support of this interpretation, the SUC2 ORF was found at the nuclear periphery in 45% of wild-type cells under repressing conditions; this is significantly above the 33.3% expected by chance, indicating that the distribution of the gene under these conditions is not random.
Importantly, perinuclear sublocalization of Mig1 is required for its function. In glucose-grown Δhxk2 cells, where it fails to repress SUC2 transcription (Neigeborn and Carlson 1984, 1987), Mig1 remains nuclear but is excluded from the perinuclear compartment (Figure 4B). These data favor reverse recruitment (Figure 5C) because they link the perinuclear sublocalization of an upstream transcriptional regulator with its function in repressing gene expression.
Figure 5.—
Models for localization of induced genes to nuclear substructures. The relocation of active genes can be modeled in three distinct ways. (A) In the processing and export model, the transcript produced by active RNA polymerase II (POL) associates with a nuclear pore complex (NPC), resulting in the relocation of the active gene to the nuclear periphery. (B) Alternatively, an early event in transcriptional activation, such as promoter opening or initiation, triggers the relocation of a gene. (C) Finally, the movement of a gene may bring it into contact with a nuclear substructure, such as the NPC, where RNA polymerase II molecules are tethered. We refer to this model as reverse recruitment. The central concept of this model is that the promoter of a gene mediates both its movement to the periphery and its interaction with nuclear substructures. (D) The reverse recruitment model can also be applied to lumenal substructures. It is possible that there exist subsets of eukaryotic genes that are brought to the nuclear periphery via each of the models in A–D.
Although a number of important questions remain, a growing body of evidence favors a model of regulated gene expression based on localization to nuclear substructures rather than on diffusion-based assembly of the transcriptional machinery. The multiple possible mechanisms depicted in Figure 5 highlight the need for a definitive analysis of the sequence of events that take place immediately following eukaryotic gene induction. Movement of induced genes to the nuclear periphery raises at least two new and fundamental questions about the nature of transcriptional regulation in eukaryotes. First, which factors both accompany active genes to the periphery and are required for this migration? These “perinuclear shuttling factor(s)” may be old friends that possess this previously unrecognized talent, or they may have thus far eluded characterization. Second, what are the structure/function relationships in factors that contribute to perinuclear gene expression? Answers to these questions should result in a more accurate and detailed view of how eukaryotic genes are turned on or off.
In combination with our earlier analysis of Rap1/Gcr1 activation, the data presented here suggest that reverse recruitment is a prevalent mechanism of gene activation in yeast cells. Analogous regulatory movement of genes to a defined physical location in the nucleus may also occur in multicellular eukaryotes. Three-dimensional fluorescence in situ hybridization (FISH) has shown that in T-lymphocytes, the q11–13 region of chromosome XV pairs with its homolog and moves to a specific spatial location. Interestingly, this association is defective in patients with either Prader–Willi syndrome or Angelman syndrome, genetic diseases characterized by cognitive impairment and developmental delay (LaSalle and Lalande 1996; Cremer and Cremer 2001). Since much of the work to date on eukaryotic transcriptional regulation is predicated upon the similarity between S. cerevisiae and multicellular organisms, it seems likely that an accurate and comprehensive depiction of eukaryotic gene regulation will require an improved understanding of nuclear ultrastructures and their reverse recruitment interactions with chromatin.
Acknowledgments
We are very grateful to Michael Rout (Rockefeller University) for the generous gift of antibodies; to Balaraj Menon for contributing QFPD data; and to Martha Sparrow, Kristina Clarke, and Baobin Kang for outstanding technical support. We also thank Jeong-Ho Kim for helpful discussions and critical review of the manuscript. This work was supported by National Institutes of Health grant RR16476 and National Science Foundation grant EPS0132618.
References
- Barbara, K. E., T. M. Haley, K. A. Willis and G. M. Santangelo, 2007. The transcription factor Gcr1 stimulates cell growth by participating in nutrient-responsive gene expression on a global level. Mol. Genet. Genomics 277: 171–188. [DOI] [PubMed] [Google Scholar]
- Boukaba, A., E. I. Georgieva, F. A. Myers, A. W. Thorne, G. Lopez-Rodas et al., 2004. A short-range gradient of histone H3 acetylation and Tup1p redistribution at the promoter of the Saccharomyces cerevisiae SUC2 gene. J. Biol. Chem. 279: 7678–7684. [DOI] [PubMed] [Google Scholar]
- Brickner, J. H., and P. Walter, 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal, G. G., A. Genovesio, S. Rodriguez-Navarro, C. Zimmer, O. Gadal et al., 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773. [DOI] [PubMed] [Google Scholar]
- Carlson, M., 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2: 202–207. [DOI] [PubMed] [Google Scholar]
- Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus et al., 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Casolari, J. M., C. R. Brown, D. A. Drubin, O. J. Rando and P. A. Silver, 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, P. R., 2001. Principles of Nuclear Structure and Function. Wiley–Liss, New York.
- Cremer, T., and C. Cremer, 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2: 292–301. [DOI] [PubMed] [Google Scholar]
- DeVit, M. J., and M. Johnston, 1999. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9: 1231–1241. [DOI] [PubMed] [Google Scholar]
- De Vit, M. J., J. A. Waddle and M. Johnston, 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8: 1603–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian, K. D., 1980. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol. Gen. Genet. 178: 633–637. [DOI] [PubMed] [Google Scholar]
- Granneman, S., and S. J. Baserga, 2005. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr. Opin. Cell Biol. 17: 281–286. [DOI] [PubMed] [Google Scholar]
- Hardie, D. G., D. Carling and M. Carlson, 1998. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67: 821–855. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Kipper, J., C. Strambio-de-Castillia, A. Suprapto and M. P. Rout, 2002. Isolation of nuclear envelope from Saccharomyces cerevisiae. Methods Enzymol. 351: 394–408. [DOI] [PubMed] [Google Scholar]
- Lamond, A. I., and J. E. Sleeman, 2003. Nuclear substructure and dynamics. Curr. Biol. 13: R825–R828. [DOI] [PubMed] [Google Scholar]
- LaSalle, J. M., and M. Lalande, 1996. Homologous association of oppositely imprinted chromosomal domains. Science 272: 725–728. [DOI] [PubMed] [Google Scholar]
- Lutfiyya, L. L., V. R. Iyer, J. DeRisi, M. J. DeVit, P. O. Brown et al., 1998. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150: 1377–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., and R. Reed, 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Menon, B. B., N. J. Sarma, S. Pasula, S. J. Deminoff, K. A. Willis et al., 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102: 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C., Z. Wang, R. G. Roeder and J. G. Gall, 2002. RNA polymerase III in cajal bodies and lampbrush chromosomes of the Xenopus oocyte nucleus. Mol. Biol. Cell 13: 3466–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn, L., and M. Carlson, 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn, L., and M. Carlson, 1987. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115: 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T., 2002. Dynamics and genome-centricity of interchromatin domains in the nucleus. Nat. Cell Biol. 4: E287–E291. [DOI] [PubMed] [Google Scholar]
- Pi, H., C. T. Chien and S. Fields, 1997. Transcriptional activation upon pheromone stimulation mediated by a small domain of Saccharomyces cerevisiae Ste12p. Mol. Cell. Biol. 17: 6410–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl, C., 1885. Uber zeillteilung. Morphol. Jahrbuch 10: 214–330. [Google Scholar]
- Sacco-Bubulya, P., and D. L. Spector, 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo, G. M., 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 253–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, H., and H. Uemura, 2005. Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast 22: 111–127. [DOI] [PubMed] [Google Scholar]
- Schmid, M., G. Arib, C. Laemmli, J. Nishikawa, T. Durussel et al., 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21: 379–391. [DOI] [PubMed] [Google Scholar]
- Schmidt, M. C., and R. R. McCartney, 2000. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19: 4936–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., M. Lutzmann, H. Santos-Rosa, K. Leonard, S. Mueller et al., 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149: 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K. P., P. T. Moen, K. L. Wydner, J. R. Coleman and J. B. Lawrence, 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., G. Van Houwe, F. Hediger, V. Kalck, F. Cubizolles et al., 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778. [DOI] [PubMed] [Google Scholar]
- Treitel, M. A., S. Kuchin and M. Carlson, 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6273–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkel, S., T. Turgut, M. C. Lopez, H. Uemura and H. V. Baker, 2003. Mutations in GCR1 affect SUC2 gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 268: 825–831. [DOI] [PubMed] [Google Scholar]
- Vallier, L. G., and M. Carlson, 1994. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics 137: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, O., R. Townley, S. Kuchin and M. Carlson, 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15: 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan and P. Hemmerich, 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., M. Pierce, L. Schneper, C. G. Guldal, X. Zhang et al., 2004. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2: 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., S. J. Deminoff and G. M. Santangelo, 1997. Specialized Rap1p/Gcr1p transcriptional activation through Gcr1p DNA contacts requires Gcr2p, as does hyperphosphorylation of Gcr1p. Genetics 147: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]