Abstract
The relationship between telomeres and nonhomologous end-joining (NHEJ) is paradoxical, as NHEJ proteins are part of the telomere cap, which serves to differentiate telomeres from DNA double-strand breaks. We explored these contradictory functions for NHEJ proteins by investigating their role in Kluyveromyces lactis telomere metabolism. The ter1-4LBsr allele of the TER1 gene resulted in the introduction of sequence altered telomeric repeats and subsequent telomere–telomere fusions (T–TFs). In this background, Lig4 and Ku80 were necessary for T–TFs to form. Nej1, essential for NHEJ at internal positions, was not. Hence, T–TF formation was mediated by an unusual NHEJ mechanism. Rad50 and mre11 strains exhibited stable short telomeres, suggesting that Rad50 and Mre11 were required for telomerase recruitment. Introduction of the ter1-4LBsr allele into these strains failed to result in telomere elongation as normally observed with the ter1-4LBsr allele. Thus, the role of Rad50 and Mre11 in the formation of T–TFs was unclear. Furthermore, rad50 and mre11 mutants had highly increased subtelomeric recombination rates, while ku80 and lig4 mutants displayed moderate increases. Ku80 mutant strains also contained extended single-stranded 3′ telomeric overhangs. We concluded that NHEJ proteins have multiple roles at telomeres, mediating fusions of mutant telomeres and ensuring end protection of normal telomeres.
TELOMERES are specialized protein–DNA complexes at the ends of linear chromosomes. Telomeric DNA in most eukaryotes consists of tandem repeats of short G-rich sequences. In Saccharomyces cerevisiae and Kluyveromyces lactis, telomeric repeats are bound by repressor activator protein 1 (Rap1) (Conrad et al. 1990; Krauskopf and Blackburn 1996), a sequence-specific DNA-binding protein containing two Myb domains. Myb-domain proteins bind to telomeres in other organisms as well, for example, Taz1 in fission yeast (Cooper et al. 1997) and Trf1/Trf2 in mammals (Broccoli et al. 1997).
Telomeres are synthesized by a specialized enzyme called telomerase (Chan and Blackburn 2004). In both S. cerevisiae and K. lactis, the catalytic core of this enzyme is composed of a reverse transcriptase, Est2, and an internal RNA component (TLC1 in S. cerevisiae and TER1 in K. lactis) that serves as template during DNA synthesis. Complete loss of telomerase causes shortening of telomeres and leads to replicative senescence due to the inability of the traditional DNA replication machinery to fully synthesize linear DNA ends (Lundblad and Szostak 1989; McEachern and Blackburn 1996).
Natural chromosome termini are protected from fusing with other DNA ends, indicating that telomeres have properties that differentiate them from DNA double-strand breaks (DSBs) elsewhere in the genome. This protective feature of telomeres, first observed by McClintock (1941) and Müller (1938), is often referred to as the telomere-cap function. One result of a compromised telomere cap is the recognition of chromosome termini as DSBs and subsequent attempts by the cell to repair these breaks. One pathway for DSB repair is the homologous recombination (HR) pathway, dependent on the RAD52 group of genes (Symington 2002). The hallmark of the HR pathway is the use of a sister chromatid or homologous chromosome as a template during a mostly error-free repair process. In contrast, the nonhomologous end-joining (NHEJ) pathway for DSB repair (Daley et al. 2005) requires little or no homology and simply fuses two free DNA ends, often generating small deletions and insertions in the process. Normally, these ends are the result of DSBs that arise at internal chromosome positions. The Ku70/Ku80 heterodimer (Milne et al. 1996) is required for NHEJ and is thought to protect and align the DNA ends for subsequent end processing and ligation (Feldmann et al. 2000). The DNA ligase, Lig4 (Dnl4 or Lig IV), (Schär et al. 1997; Teo and Jackson 1997; Wilson et al. 1997) and Lif1 (Lig IV interactingfactor 1) (Herrmann et al. 1998) provide the ligase activity during NHEJ. Lif1 is the yeast functional homolog of mammalian Xrcc4 and is required for the stability and full activity of Lig4 (Teo and Jackson 2000). In addition, the MRX complex (Mre11, Rad50, and Xrs2) is also required for efficient NHEJ in S .cerevisiae and K. lactis (Moore and Haber 1996; Boulton and Jackson 1998; Kegel et al. 2006). Mre11 is an endo/exo nuclease, but mutant forms of Mre11 lacking detectable nuclease activity still support NHEJ (Lewis et al. 2004). Cell type regulates NHEJ in S. cerevisiae, such that haploid MATa or MATα strains perform NHEJ efficiently, while diploid MATa/MATα strains perform NHEJ inefficiently (Åström et al. 1999; Lee et al. 1999). This regulation of NHEJ is the result of transcriptional repression of the NEJ1 gene in diploid cells. NEJ1 encodes a protein that interacts with Lif1 and is essential for NHEJ by all standard assays (Frank-Vaillant and Marcand 2001; Kegel et al. 2001; Ooi et al. 2001; Valencia et al. 2001). The K. lactis NEJ1 ortholog is also essential for NHEJ, but its transcription is not regulated by cell type, and K. lactis diploid cells perform NHEJ with an efficiency similar to either haploid cell type (Kegel et al. 2006). In humans, the recently identified XLF/Cernunnos gene was suggested to be the ortholog of yeast NEJ1 (Callebaut et al. 2006).
It is known that telomeres with a compromised cap can engage in both HR and NHEJ (Lundblad and Blackburn 1993; McEachern and Blackburn 1996). HR repair can result in telomerase-independent telomere elongation or shortening. In contrast, NHEJ of telomeres results in telomere–telomere fusions (T–TFs). This is illustrated in telomerase-deficient fission yeast in which the majority of cells die as a result of gradual telomere attrition. In most surviving cells, chromosomes lacking detectable telomeric sequence circularize as a result of T–TFs, while a smaller fraction of survivors appear to maintain their telomeres by recombination (Nakamura et al. 1998). In addition, in S. cerevisiae and fission yeast, both telomerase and Tel1, a member of the ATM family of protein kinases, protect telomeres from fusions (Nakamura et al. 1998; Chan and Blackburn 2003; Mieczkowski et al. 2003).
Other mutations related to telomere function can also compromise the telomere cap. For example, overexpression of a dominant-negative form of the human telomere protein hTrf2 resulted in T–TFs in which telomere sequences were still present (van Steensel et al. 1998). Fission yeast Taz1 (Ferreira and Cooper 2001) and S. cerevisiae Rap1 (Pardo and Marcand 2005) also protect telomeres from fusing, suggesting that these Myb-domain proteins are part of the DNA–protein complex constituting the cap. In K. lactis, T–TFs are observed in strains with specific mutations in the template region of the telomerase RNA. These mutations are predicted to abolish or decrease Rap1 binding to the telomere (McEachern et al. 2000). T–TFs containing telomeric repeats appear to arise by a NHEJ-dependent mechanism since their formation requires Lig4 in both yeasts and mammals (Smogorzewska et al. 2002; Liti and Louis 2003; Mieczkowski et al. 2003; Ferreira and Cooper 2004). The role of individual NHEJ components in generating T–TFs is not clear, however. S. cerevisiae Ku has been reported to be both required (Pardo and Marcand 2005) and dispensable (Mieczkowski et al. 2003) for the formation of T–TFs. Paradoxically, knockdown alterations of mammalian Ku86 expression promote chromosome fusions involving telomeric sequences (Hsu et al. 2000; Samper et al. 2000), suggesting that Ku acts to prevent formation of T–TFs. In addition, S. cerevisiae Nej1 has been reported to prevent T–TFs, despite being required for promoting NHEJ (Liti and Louis 2003).
In this study, we explored the role of NHEJ proteins in telomere metabolism in K. lactis. We found that Ku80 and Lig4 were required for the formation of T–TFs and that subtelomeric gene conversion rates were dramatically increased in strains lacking Mre11 or Rad50. Surprisingly, Nej1 was not required for T–TF formation.
MATERIALS AND METHODS
Yeast strains:
The strains used in this study are listed in Table 1. Strains lacking LIG4 (AKY124), NEJ1 (SAY572), KU80 (SAY573), MRE11 (SAY559), and RAD50 (SAY557) were generated previously (Kegel et al. 2006). The double-mutant strains sir4 lig4 (SAY703), sir4 nej1 (SAY701), sir4 ku80 (AKY116), and sir4 mre11 (SAY704) were generated by crossing a sir4 strain (SAY100) with the respective nonhomologous-end-joining mutant strain followed by tetrad analysis. For T–TF assays, a strain containing a duplicated TER1 locus containing both a wild-type and the mutant ter1-4L Bsr allele (generated by plasmid loop-in) was crossed to strains carrying null mutations in LIG4, NEJ1, KU80, MRE11, and RAD50. Tetrad analysis resulted in the isolation of TER1∷URA3-ter1-4L-Bsr ku80 (SAY600), TER1∷URA3-ter1-4L Bsr mre11 (SAY579), TER1∷URA3-ter1-4L Bsr rad50 (SAY604), TER1∷URA3-ter1-4L Bsr nej1 (SAY562), and TER1∷URA3-ter1-4L Bsr lig4 (SAY563) double-mutant strains. The TER1∷URA3-ter1-4LBsr nej1 lig4 triple-mutant strain (SAY564) was obtained through the same procedure following a cross between 4LBsr and a nej1 lig4 double-mutant strain (SAY545). Additionally, TER1∷URA3-ter1-4L Bsr single-mutant strains were isolated from each of the above-mentioned crosses to serve as controls. Each strain was subsequently plated onto 5-FOA medium (Boeke et al. 1987) and clones that had lost the wild-type TER1 gene but retained the mutant ter1-4L Bsr allele were identified. The resulting strains were maintained by serial restreaks on rich medium (YEPD) for 48–72 hr at 30°.
TABLE 1.
Yeast strains used in this study
| K. lactis | Genotype | Reference |
|---|---|---|
| 4LBsr | MATα his2-2 trp1 ura3-1 TER1∷URA3-ter1-4LBsr | McEachern et al. (2000) |
| CK213-4C | MATaleu2 lysA1 metA1 trp1 uraA1 | Chen and Clark-Walker (1994) |
| AccSna | MATahis2-2 trp1 uraA1 ter1-AccSna | McEachern et al. (2000) |
| AKY116 | MATalysA1 uraA1 ku80∷LEU2 sir4∷LEU2 | This study |
| AKY124 | MATaleu2 lysA1 trp1 uraA1 lig4∷KANMX | Kegel et al. (2006) |
| SAY100 | MATα ade2 leu2 uraA1 sir4∷LEU2 | Åström and Rine (1998) |
| SAY545 | MATaleu2 lysA1 metA1 trp1 uraA1 nej1∷LEU2 lig4∷KANMX | This study |
| SAY557 | MATaleu2 lysA1 metA1 trp1 uraA1 rad50∷KANMX | Kegel et al. (2006) |
| SAY559 | MATaleu2 lysA1 metA1 trp1 uraA1 mre11∷KANMX | Kegel et al. (2006) |
| SAY561 | MATα his2-2 leu2 uraA1 trp1 TER1∷URA3-ter1-4LBsr | This study |
| SAY562 | MATalysA1 or LYSA1trp1uraA1 nej1∷LEU2 TER1∷URA3-ter1-4LBsr | This study |
| SAY563 | MATalysA1 or LYSA1 leu2 uraA1 trp1 lig4∷KANMX TER1∷URA3-ter1-4LBsr | This study |
| SAY564 | MAT? his2-2 lysA1 or LYSA1 trp1 uraA1 lig4∷KANMX nej1∷LEU2 TER1∷URA3-ter1-4LBsr | This study |
| SAY572 | MATaleu2 lysA1 metA1 trp1 uraA1 nej1∷LEU2 | Kegel et al. (2006) |
| SAY573 | CK213-4C ku80∷LEU2 | Kegel et al. (2006) |
| SAY579 | MATahis2-2 metA1 uraA1 mre11∷KANMX TER1∷URA3-ter1-4LBsr | This study |
| SAY580 | MATα his2-2 uraA1 TER1∷URA3-ter1-4LBsr | This study |
| SAY600 | MATα uraA1 ku80∷LEU2 TER1∷URA3-ter1-4LBsr | This study |
| SAY604 | MATahis2-2 leu2 metA1 uraA1 rad50∷KANMX TER1∷URA3-ter1-4LBsr | This study |
| SAY605 | MATα his2-2 TER1∷URA3-ter1-4LBsr | This study |
| SAY606 | MATaleu2 lysA1 uraA1 rad50∷KANMX TER1∷URA3-ter1-4LBsr | This study |
| SAY701 | MATα leu2 metA1 uraA1 nej1∷LEU2 sir4∷LEU2 | This study |
| SAY703 | MATα ade1 leu2 lysA1 metA1 trp1 uraA1 lig4∷KANMX sir4∷LEU2 | This study |
| SAY704 | MATaleu2 lysA1 uraA1 mre11∷KANMX sir4∷LEU2 | This study |
| Kl7B520 | MATahis2-2 trp1 uraA1 | Wray et al. (1987) |
| GG1958 | MATα ade2 | This laboratory |
Random spore analysis:
The ter1-AccSna strain used in this experiment is a derivative of the wild-type haploid strain 7B520 (ura3-1 his2-2 trp1) (Wray et al. 1987). Analysis was performed on the spores generated by mating the ter1-AccSna strain with GG1958 (ade2). Diploid cells were plated onto sporulation media for 4 days. Cells were then incubated in 200 μ1 of zymolyase (0.17 mg/ml) in 1 m sorbitol at 37° for 10 min to digest the asci. Subsequently, cells were subjected to heat treatment at 55° for 10 min to kill all vegetative cells. The resulting spores were serially diluted in TE (10 mm Tris and 1 mm EDTA) and plated onto YPD plates.
Hybridizations:
DNA blots were hybridized with the previously described wild-type K. lactis telomeric probe (Klac1-25) at 50° following a standard protocol (McEachern and Blackburn 1995). In-gel hybridization experiments were performed as previously described (Dionne and Wellinger 1996), using oligonucleotide probes complementary to either the 5′ telomeric strand (G-probe) (Klac1-25) or the 3′ telomeric strand (C-probe). Approximately 3 μg of digested DNA was electrophoresed through a 0.7% agarose gel and analyzed using the conditions described in Underwood et al. (2004).
Subtelomere gene conversion assay:
The gene conversion assay was performed according to the protocols described previously (McEachern and Iyer 2001). In brief, a native telomere in rad50, mre11, ku80, lig4, and wild-type strains was replaced by transformation with an ∼2.0-kb EcoRI and SacII subtelomeric URA3 (STU) fragment containing the URA3 gene from S. cerevisiae. The marker was inserted ∼120 bp from the junction of the cloned telomere and subtelomere fragments of K. lactis. Serially diluted cells of the clones containing a single-copy STU fragment were plated onto synthetic complete (SC) medium, SC + 5-FOA, and YPD. Measurement of the loss of the URA3 gene was performed by counting colonies grown on 5-FOA and comparing them to the total number of colonies grown on YPD and SC.
Analysis of telomere–telomere fusions:
The strains used for analysis of T–TFs were ter1-4LBsr strains SAY605 and SAY561 following 25 and 5 serial restreaks, respectively. Genomic DNA was prepared using the MasterPure Yeast DNA purification kit (Epicentre, Madison, WI). One primer (P1 5′-CAGGGCGGGTAACATGAC-3′) contained a sequence unique to K. lactis chromosome 2L and corresponded to 268–285 nucleotides upstream from the telomeric repeat sequence start (Nickles and McEachern 2004). The second primer (P2-11 5′ GAAAGAGGAAATCCGTTTCG-3′) contained a sequence common to a subtelomeric region of at least nine chromosome ends other than 2L. Potential annealing sites for this primer ranged between 167 and 254 nucleotides from the telomeric repeat sequence start, depending upon the chromosome end (Nickles and McEachern 2004). PCR reactions (50 μl) contained 0.2 ng genomic DNA, 1× PCR reaction buffer (10 mm Tris–HCl, 1.5 mm MgCl2, 50 mm KCl), dNTP 0.1 mm each, primers 0.5 μm each, and 1 unit Taq DNA polymerase. Reaction conditions were as follows: 95° for 1 min; 35 cycles of 95° for 30 sec, 55° for 30 sec, 72° for 2 min, followed by 72° for 10 min. The products were separated on a 1% agarose gel and visualized by ethidium bromide (EtBr) staining. Portions of the resulting DNA smear were cut from the gel and the DNA products were recovered. Isolated products were then cloned into the Promega (Madison, WI) pGEM T-Easy vector using the suggested protocol and transformed into the Escherichia coli strain DH5α cells. Approximately 1 μg of plasmid DNA was digested with EcoRI or doubly digested with EcoRI and BsrGI. The mixtures were separated on a 2% agarose gel and visualized by ethidium bromide staining to determine fragment sizes. Clones were submitted for DNA sequencing by Macrogen (Seoul, South Korea).
RESULTS
Telomere lengths in strains lacking NHEJ components:
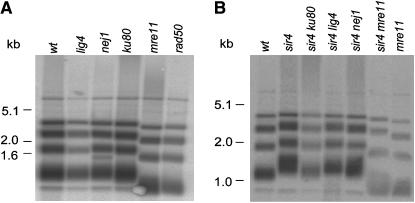
To explore if NHEJ proteins were required for maintaining normal telomere length in K. lactis, we determined telomere lengths in strains lacking individual NHEJ proteins. The 12 telomeres in K. lactis give rise to a characteristic pattern of four to five bands on DNA blots after digestion of the genomic DNA with EcoRI (Figure 1A). Comparing the pattern using DNA from strains with null alleles in NHEJ genes revealed that lig4, ku80, and nej1 mutant strains had normal telomere lengths, but telomeres were shorter in rad50 and mre11 mutants. The shortened telomeres in strains lacking the MRX complex were consistent with previous results obtained in S. cerevisiae (Tsukamoto et al. 2001). The observation that a ku80 mutant strain had normal length telomeres was surprising since the corresponding mutation leads to telomere shortening in S. cerevisiae.
Figure 1.—
Telomere length in strains compromised for NHEJ. DNA blot of EcoRI-digested chromosomal DNA hybridized with a probe specific to K. lactis telomeric repeats. (A) Telomere lengths from strains CK213-4C (wt, or wild type), AKY124 (lig4), SAY572 (nej1), SAY573 (ku80), SAY559 (mre11), and SAY557 (rad50). (B) Telomere lengths of strains SAY100 (sir4), AKY116 (sir4 ku80), SAY703 (sir4 lig4), SAY701 (sir4 nej1), SAY704 (sir4 mre11), and SAY559 (mre11). Molecular weight markers are in kilobases.
Next we investigated if we could observe a telomere length defect in the absence of NHEJ proteins in a sir4 background. It had previously been shown that strains lacking Sir4 in K. lactis have longer telomeres than wild-type strains (Åström and Rine 1998). We hypothesized that using a sir4 mutant background might sensitize the assay. The telomere lengths of a sir4 single and a sir4 ku80 double-mutant strain indeed differed. The double mutant had telomeres longer than the wild-type strain, but shorter than the sir4 single-mutant strain (Figure 1B). In contrast, telomere lengths in the nej1 sir4 and lig4 sir4 double-mutant strains were similar to the sir4 single-mutant strain, confirming that NHEJ function was not required for maintaining normal telomere length (Figure 1B). Telomere length in the mre11 sir4 double-mutant strain was shorter than that of the wild-type strain, but longer than that of the mre11 single mutant, indicating that Mre11 and Sir4 affected telomere length homeostasis by different and opposing mechanisms.
Single-stranded 3′ overhangs were formed in the ku80 mutant:
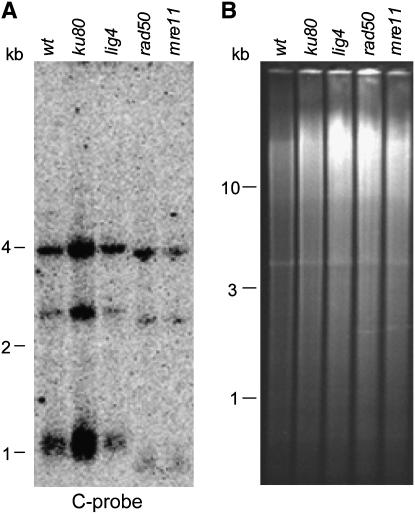
One manifestation of telomere capping loss is the generation of extended single-stranded 3′ telomeric overhangs. We therefore employed nondenaturing in-gel hybridization, a method that detects relative amounts of 3′ overhangs, to analyze ku80, lig4, rad50, and mre11 mutant strains (Dionne and Wellinger 1996). Using a probe complementary to the 3′ strand (C-probe), a strong signal was visible in the ku80 strain relative to wild type, while only weak signals were detected in the lig4, mre11, and rad50 mutant strains (Figure 2A). No signal was observed using a probe complementary to the 5′ telomeric DNA (G-probe) (data not shown). EtBr staining of the gel prior to hybridization confirmed that similar amounts of DNA were loaded in all lanes (Figure 2B). These results showed that extended 3′ overhangs were present in the ku80 strain and are consistent with Ku80 contributing to telomere end protection. The precise length of these overhangs and whether they were cell cycle dependent remains unclear.
Figure 2.—
Long 3′ overhangs in ku80 cells. Nondenaturing in-gel hybridization of genomic DNA digested with PstI from wt (wild type), ku80, lig4, mre11, and rad50 strains. (A) Shows hybridization to a G-strand telomeric probe. (B) Shows the EtBr-stained gel prior to hybridization. Molecular weight markers are in kilobases.
Increased subtelomeric recombination in rad50, mre11, lig4, and ku80 mutants:
Both shorter telomere length and elongated 3′ telomere overhangs have been associated with increased recombination in and near telomeres (McEachern and Iyer 2001; Underwood et al. 2004; Iyer et al. 2005). We therefore hypothesized that ku80, mre11, and rad50 mutant strains would exhibit elevated levels of subtelomeric recombination. To test this idea, we performed assays on ku80, mre11, rad50, lig4, and wild-type strains to measure the rates of subtelomeric gene conversion as previously described (McEachern and Iyer 2001). This assay measures the loss of a URA3 marker gene, which was artificially positioned close to a telomere in each of the different strains (see materials and methods). Silencing of the URA3 gene is not observed using these constructs since 108/108 5-FOA resistant clones derived from cells with the same URA3-tagged telomere had all lost the URA3 gene as judged by DNA blots (Natarajan et al. 2006). Relative to the wild-type control strain, we observed dramatic increases in URA3 loss (Table 2). The ∼250- to 1000-fold increased gene conversion rates in rad50 and mre11 strains indicate that these mutants were particularly compromised in their ability to protect telomeric and subtelomeric regions against recombination. An interesting observation is the modest increase in subtelomeric recombination in the absence of Lig4. This is the first observation that Lig4 might have a role in telomere end protection. We confirmed that the URA3 gene had been lost from 5-FOA-resistant clones from the lig4 and ku80 mutant strains using DNA blots (data not shown). Our results indicate that despite possessing telomeres of normal length, ku80 and lig4 mutants have a telomere-capping defect.
TABLE 2.
Elevated levels of recombination near telomeres in rad50, mre11, ku80, and lig4 mutant strains
| Strains | Gene conversion rates [mutation rate ± SE (n)] | Relative rate |
|---|---|---|
| Wild type | 2.8 × 10−6 ± 1.5 × 10−6 (13) | 1 |
| ku80 | 2.3 × 10−5 ± 9.5 × 10−6 (15) | ∼8 |
| lig4 | 3.5 × 10−5 ± 6.0 × 10−6 (10) | ∼13 |
| rad50 | 6.5 × 10−4 ± 8.8 × 10−5 (18) | ∼230 |
| mre11 | 2.8 × 10−3 ± 1.1 × 10−3 (15) | ∼1000 |
The gene conversion rates of the mutants are based on measurement of several (three to four) independent transformants of the individual mutant strains, containing the URA3 gene inserted near one telomere (see materials and methods). For each strain, the number of samples, n, used for the assay is in parentheses. The standard error (SE) was calculated as the standard deviation divided by the square root of n.
Telomere–telomere fusions were produced by an atypical NHEJ mechanism:
Mutations in the template region of telomerase RNA, encoded by the K. lactis TER1 gene, lead to the introduction of the corresponding changes in newly synthesized telomeric repeats (McEachern and Blackburn 1995; Underwood et al. 2004). Specific mutations in the TER1 gene, predicted to abolish Rap1 binding to the newly synthesized repeats, can lead to the formation of T–TFs (McEachern et al. 2000). We used strains containing a combination of a particular TER1 allele, termed ter1-4LBsr (abbreviated ter1-4L), with mutations in NHEJ components and asked if T–TFs were formed.
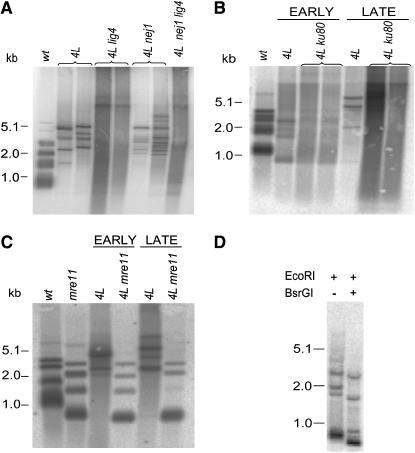
The ter1-4L single-mutant strains used as controls showed the expected pattern of immediate severe telomere elongation and degradation, as indicated by a smeared DNA blot signal. After prolonged passaging, the smeared signal was resolved into a series of sharp, discrete bands (Figure 3B, 4L early vs. late). These bands were previously shown to be indicative of T–TFs (McEachern et al. 2000). Two independent isolates of ter1-4L strains lacking Lig4 or Ku80 showed no signs of T–TFs even after 10 and 25 streaks, respectively (Figure 3, A and B). In contrast, strains lacking Nej1 formed T–TFs at a rate similar to those of a wild-type strain (Figure 3A and data not shown). We concluded that K. lactis T–TFs induced by the ter1-4L allele appeared to arise by an atypical NHEJ mechanism, a mechanism that required Lig4 and Ku80, but not Nej1.
Figure 3.—
T–TFs in NHEJ mutant strains. (A) LIG4, but not NEJ1, was required for formation of T–TFs. Shown is a DNA blot using two independent isolates of ter1-4LBsr (ter1-4L), ter1-4L lig4, and ter1-4L nej1 and one ter1-4L lig4 nej1 strain. Chromosomal DNA was prepared from the strains following 10 streaks. (B) KU80 was required for formation of T–TFs. The blot is divided into early and late lanes. Chromosomal DNA from early lanes was isolated immediately following the generation of the mutant strains. Late lanes are of the same mutant strains following 24 serial restreaks. Two independent isolates of ter1-4L ku80 and one isolate of a ter1-4L single mutant are shown. (C) ter1-4L mre11 double-mutant strains had short, stable telomeres. The blot is divided into early and late lanes as described above. Shown are ter1-4L, mre11, and ter1-4L mre11 strains. Wild types for all blots are TER1 strains recovered from the same tetrads used in the generation of the respective mutant strains. (D) A ter1-4L rad50 double-mutant strain incorporated mutant repeats. EcoRI and EcoRI/BsrGI digestions of genomic DNA isolated from a ter1-4L rad50 mutant strain following 24 serial restreaks. Molecular weight markers are in kilobases.
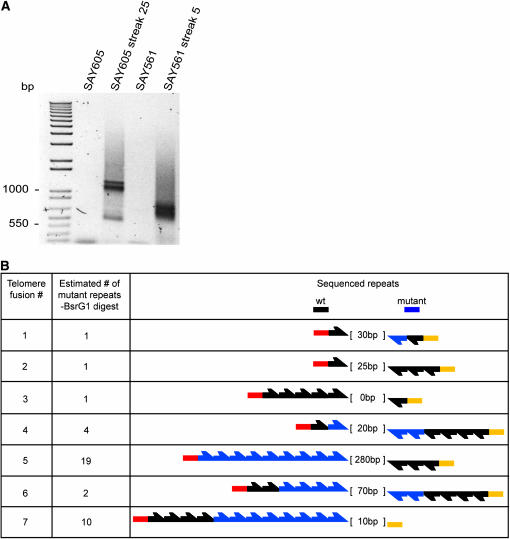
Figure 4.—
Molecular characterization of T–TFs. (A) Agarose gel electrophoresis of PCR amplifications using genomic DNA from pre- and postfusion strains, as indicated above the lanes, and subtelomeric specific primers. Results were from strains SAY561 and SAY605 following 5 and 25 serial restreaks, respectively. Control lanes result from PCR of genomic DNA isolated from the parental strains used in the generation of each mutant strain prior to the TER1 loop-out procedure. Size markers indicated on the left. (B) K. lactis T–TFs analyzed by restriction digests and DNA sequencing. Seven different T–TFs (first column) were analyzed, one to three originating from strain SAY605 and four to seven from strain SAY561. The second column shows an estimate of the number of mutant repeats present as determined by BsrGI digestion of each cloned fragment followed by agarose gel electrophoresis. The third column is a schematic of the sequencing results. Sequenced wild-type repeats (black arrows) and mutant repeats (blue arrows) are indicated. Red and yellow boxes represent subtelomeric sequences. The numbers within brackets indicate the estimated difference, in base pairs, between the fragment submitted for sequencing and the actual sequence recovered from the analysis.
Combining the ter1-4L mutation with rad50 or mre11 mutations revealed a surprising result with respect to the initial telomere elongation phenotype. The ter1-4L rad50 and the ter1-4L mre11 double-mutant strains did not show a loss of telomere length regulation and the telomeres remained stably short even after 25 streaks (Figure 3C and data not shown). In addition, we observed no signs of T–TFs in either strain. A useful feature of this particular TER1 mutant allele is that it introduces a BsrGI restriction site into each newly synthesized telomeric repeat. Digestion with BsrGI showed that in spite of failing to undergo telomere elongation, a few mutant repeats had indeed been incorporated in the rad50 and mre11 mutant strains (Figure 3D and data not shown). Hence, these results were consistent with a role for Mre11 and Rad50 in telomerase recruitment, but the contribution of these proteins to the formation of T–TFs remains unclear.
Molecular characterization of telomere–telomere fusions:
The fusions between telomeres were further characterized using a PCR-based assay similarly to a previously described strategy in S. cerevisiae (Mieczkowski et al. 2003; Pardo and Marcand 2005). Taking advantage of recent sequence data (Nickles and McEachern 2004), we designed four PCR primers that annealed within the subtelomeric regions of K. lactis telomeres. We tested the specificity of these primers using chromosomal DNA from nonfused and postfusion strains as template. One primer pair specifically and consistently amplified DNA from several postfusion strains (Figure 4A), resulting in a smeared signal. This pair corresponded to a primer complementary to a sequence unique to the left end of chromosome 2 (P1) and a primer that was complementary to at least nine subtelomeric regions (P2-11) (see materials and methods for details). PCR with only primer P2-11 failed to generate a product. This result did not rule out fusions between telomeres not involving telomere 2L, since such fusions, being almost perfect palindromes, probably resist PCR amplification.
The PCR products from two postfusion strains were cloned and amplified in E. coli. Digestion with BsrGI enabled us to make estimations of the number of telomeric repeats containing BsrGI sites. In addition, DNA sequencing made it possible to count most wild-type repeats. The template DNA used originated from independently isolated ter1-4L single-mutant strains from late (25) and early (5) streaks. Given the smeary nature of the signal, three or four independent clones were sequenced. In all cases, the anticipated subtelomeric sequences as well as basal telomeric repeats were identified, but the T–TFs could not be sequenced through the fusion point. The three clones originating from the late streak (Figure 4B, telomere fusions 1–3) had very few BsrGI-containing repeats, while the four clones originating from the early streak contained numerous mutant repeats (4–19). In all of the fusions sequenced, the number of wild-type repeats on each end was very limited (0–5).
We assume that the PCR-based assay for T–TFs was much more sensitive than the DNA blots used previously. Consistent with this assumption, we found that T–TFs could be detected earlier using the PCR-based assay, in fact, immediately following the pop-out procedure. Therefore, we investigated if T–TFs could be detected in the NHEJ mutant strains using the PCR-based assay. The results showed evidence of T–TFs in ter1-4L nej1 mutant strains, but not in ter1-4L strains lacking Ku80, Lig4, Mre11, or Rad50 (data not shown). The two assays used for detection of T–TFs generated the same results and were thus confirmatory.
Multiple T–TFs are highly detrimental to K. lactis meiosis:
The ter1 template mutants that cause fusions between most or all telomeres have been postulated to select for derivatives that have circularized each of their six individual chromosomes (McEachern et al. 2000). Mutant cells with no detectable free telomeres and six telomere fusions displayed only moderately slow mitotic growth. Nevertheless, such cells were expected to incur difficulties while undergoing meiosis. This is because crossovers involving a circular chromosome would dimerize a homologous chromosomal pair and interfere with their disjunction at meiosis I. To test this notion, we used a ter1-AccSna mutant for conducting mating and sporulation analysis. This mutant strain, which contains a single base substitution in both the left half and the right half of the Rap1-binding site (Figure 5), was slower to undergo fusions than the ter1-4L mutant strain. However, the T–TFs formed in this strain were more stable, presumably because of the smaller average size of the fused telomere sequences.
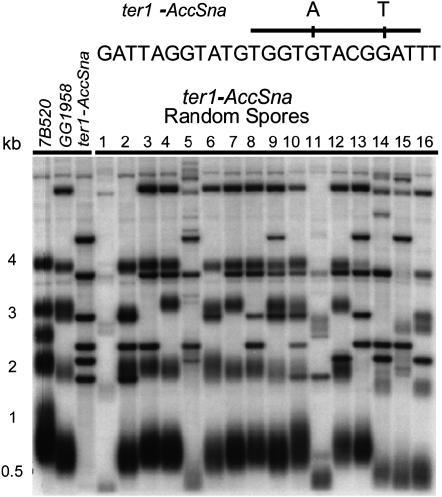
Figure 5.—
Multiple T–TFs are detrimental to the ter1-AccSna strain undergoing meiosis. Shown is a DNA blot of EcoRI-digested genomic DNA from 16 haploid strains (lanes 1–16) derived from a cross between a ter1-AccSna strain with stable T–TFs and the wild-type strain GG1958. Lanes marked as 7B520, GG1958, and ter1-AccSna represent the parental haploid controls used to set up the crosses. The sharp bands visible in lanes 1–16, which are identical in size to bands present in the ter1-AccSna parent, represent T–TFs passed unaltered through meiosis. The genomic DNA was hybridized to a G-strand telomere probe (Klac1-25). Molecular weight markers are in kilobases. Above the blot is a schematic of the ter1-AccSna allele, indicating the position of the introduced mutations.
A ter1-AccSna fusion strain showed no defect in the ability to mate with GG1958, a strain with normal telomeres, when compared to an isogenic wild-type control (7B520). However, when diploids generated by crossing wild-type 7B520 with GG1958 and ter1-AccSna with GG1958 were sporulated, a dramatic difference in successful tetrad production was observed. Microscopic observation showed that tetrads were abundant among cells of the control strain but seemingly absent from the diploids derived from the telomere fusion strain. To further characterize this meiotic defect, random spore analysis was performed on the sporulating cultures of the same diploid strains. Recovery of viable spores, following heat treatment at 55°, was consistently reduced by ∼100-fold for cells derived from the ter1-AccSna diploids relative to the control. Those cells that did survive exhibited one or more phenotypes (Ade−, Ura−, His−), which suggested that they were derivatives from the same haploid parental strains that were used to set up the initial cross. We next examined the structure of the telomeres in 16 of these segregants from the ter1-AccSna fusion diploids by DNA blots. The results confirmed that each of the clones contained mixtures of fused and unfused telomeres (Figure 5). The unfused telomeres were all either of normal length or shorter than normal, consistent with the presence of either wild-type TER1 or ter1-AccSna (which produces short telomeres prior to fusion formation). In most cases, the fusion bands were identical in size with those of the ter1-AccSna haploid parent, consistent with the fusion bands being unaltered by meiosis. In at least two instances (Figure 5, lanes 14 and 15), novel sharp bands were also visible in ter1-AccSna haploids.
DISCUSSION
We examined the role of several NHEJ genes in telomere metabolism in K. lactis. The impetus behind our work is the seemingly contradictory roles of NHEJ proteins at telomeres vs. at other genomic locations. At telomeres, several NHEJ proteins have been implicated as integral components of the protective telomere cap. In this capacity, NHEJ proteins contribute to the prevention of the very same processes that they promote within the genome. We explored several aspects of telomere maintenance, including length homeostasis, the extent of 3′ overhangs, subtelomeric gene conversion rates, and formation of T–TFs. In addition, we examined the ability of strains containing T–TFs to undergo meiosis. Taken together, this study shows that NHEJ proteins have multiple and complex roles in imparting normal telomere metabolism.
With respect to telomere length, the short telomeres of the mre11 and rad50 strains were similar to results seen in S. cerevisiae, where unusually short but stable telomeres have been seen upon deletion of certain genes in DNA repair pathways, including the MRX complex and the Tel1 kinase (Kironmai and Muniyappa 1997; Boulton and Jackson 1998). Our data argue that the K. lactis MRX complex functions in maintenance of telomere length in much the same way that it does at S. cerevisiae telomeres. In contrast, both the ku80 and sir4 mutant strains showed telomere length phenotypes different from the corresponding S. cerevisiae mutant strains. The modest telomere elongation observed in a K. lactis sir4 mutant differs from the result observed in S. cerevisiae, where both sir3 and sir4 mutations produce modest telomere shortening (Palladino et al. 1993). However, our results are consistent with the Sir4 protein being present at K. lactis telomeres and contributing in a minor way to telomere length regulation.
The Ku70/Ku80 heterodimer has been shown to be involved in both telomere length regulation and telomere capping in S. cerevisiae (Boulton and Jackson 1998; Gravel et al. 1998; Featherstone and Jackson 1999; Grandin et al. 2000; Bertuch and Lundblad 2003a,b; Stellwagen et al. 2003). Genetic analysis has shown that MRX and Tel1 function in the same pathway of telomere maintenance but in a different pathway than Ku (Ritchie and Petes 2000). Loss of either Ku70 or Ku80 caused a large decrease in telomere length and an increase in the length of 3′ overhangs present at telomeres (Bertuch and Lundblad 2003a) due to degradation of the 5′ strand of telomeric DNA (Gravel et al. 1998; Polotnianka et al. 1998; Maringele and Lydall 2002). These two roles are genetically separable (Bertuch and Lundblad 2003b). In S. cerevisiae, Ku's effect on telomere length appears to result from its ability to bind directly to a particular stem loop region of the telomerase RNA, thereby assisting in telomerase recruitment (Peterson et al. 2001; Miyoshi et al. 2003; Stellwagen et al. 2003). In this study we found that loss of the K. lactis KU80 gene does not appreciably alter telomere length. A possible explanation for this result is that the binding site for Ku at the stem loop region is absent in the telomerase RNA of K. lactis (Tzfati et al. 2003). As a consequence, the K. lactis Ku protein may not have a major role in recruitment of telomerase to telomeres. However, the observation that a ku80 sir4 double mutant exhibited slightly shorter telomeres than a sir4 single mutant is consistent with Ku contributing to telomere length regulation to some degree. Although a Ku deficiency leads to telomere shortening in many organisms studied to date (Fisher and Zakian 2005), there are exceptions to this rule. For example, loss of Ku in Arabidopsis results in considerable telomerase-dependent telomere elongation (Riha and Shippen 2003). An increased length of telomere 3′ overhangs was shared between S. cerevisiae and K. lactis ku80 mutants, indicating that the role of Ku in protecting telomeres from extensive degradation is conserved.
Large increases in subtelomeric gene conversion were previously seen in K. lactis telomerase RNA gene (TER1) mutants with stably shortened telomeres (McEachern and Iyer 2001). These events may represent break-induced replication (BIR) as subtelomeric loss of URA3 is accompanied by replacement of tagged telomeric repeats (Natarajan et al. 2006). The increased subtelomeric BIR rates seen in mre11 and rad50 mutants may therefore be an indirect consequence of shorter telomere length rather than an additional separate defect in telomere capping. A previous report found that S. cerevisiae mre11 and rad50 mutants did not display a large increase in the rate of telomeric recombination despite having very short telomeres (DuBois et al. 2002). However, this study employed a telomere capture assay that required recombination with a telomeric repeat tract that was only ∼80 bp in length. Subsequent work has shown that recombination involving homologous sequences of <100 bp can require RAD50 (Ira and Haber 2002). We hypothesize that a Rad51 filament formed on a single-stranded 3′ telomeric end could invade another telomere with the invasion extending into subtelomeric sequences. Certain outcomes of this strand invasion complex could result in loss of the URA3 gene and replacement of the original telomeric repeats with a sequence copied from the donor molecule. As the subtelomeric sequence that is shared between at least 11 of 12 K. lactis telomeres extends for well over 1 kb, there would be ample homology available for Rad51-dependent recombination to occur, even if the adjoining telomeric repeat tract was very short. Additionally, Ku deficiency has been reported to trigger increased recombination (Polotnianka et al. 1998; Baumann et al. 2002; DuBois et al. 2002; Miyoshi et al. 2003). Our results are consistent with these observations as we show that loss of ku80 results in a detectable, although moderate, increase in subtelomeric recombination.
Lig4 is an essential part of the NHEJ pathway along with Ku80, but no role has yet been assigned to Lig4 in telomere metabolism. Therefore, the 13-fold increase in recombination near telomeres in a K. lactis lig4 mutant observed in this study provides the first suggestion of Lig4 contributing to chromosome end protection at the telomeres. Alternatively, HR and NHEJ compete for repairing DSBs in subtelomeric regions and the absence of Lig4 shuttles more repair events into the HR pathway. In support of this idea, we have observed competition between HR and NHEJ for an ectopic DSB induced ∼25 kb from a telomere in K. lactis (P. Martinez and S. U. Åström, unpublished observation). It will be worthwhile to conduct further experiments to ascertain if Lig4 plays a role in telomere maintenance in the absence of telomerase.
On rare occasions, chromosome circularization has been observed in the genomes of yeasts and mammals as a result of a compromised telomere cap (Nakamura et al. 1998; Iyer et al. 2005). Several lines of evidence have pointed out that the genetic requirements for the formation of T–TFs can differ between species. In addition to the concomitant loss of all telomeric sequence, the circularized chromosomes of telomerase-deficient fission yeast have been shown to form independently of Ku80 and Lig IV (Nakamura et al. 1998; Fisher and Zakian 2005). In contrast, the T–TFs of Schizosaccharomyces pombe mutants lacking Taz1 formed between long telomeres and were dependent on Ku80 and Lig IV (Ferreira and Cooper 2001). In this study, the essential requirement of Ku80 and Lig4 provides the first evidence that fusions in the ter1-4L mutant were dependent on the NHEJ pathway. In addition, deleting the RAD50 and MRE11 genes, crucial for NHEJ at internal DSBs, led to very short telomeres and lack of any T–TFs. The simplest explanation for this result was that Rad50 and Mre11 were required for both telomere length homeostasis and for the formation of T–TFs. Another possibility could be that a certain minimum number of mutant telomeric repeats were required for T–TF formation and that this threshold was not reached in the mre11 and rad50 strains. This explanation was suggested for a T–TF-free ter1 template mutant that contained two sets of base changes that, when present in cells individually, did cause T–TFs (McEachern et al. 2000).
A key finding from this body of work was that strains lacking Nej1, a protein necessary for NHEJ at internal DSBs, formed fusions with similar rates compared to the wild type. One possibility is that the function of Nej1 was replaced by another protein during the formation of T–TFs. It was also possible that the nature of NHEJ at telomere termini differs from NHEJ at internal chromosome positions in such a manner as to render Nej1 function dispensable. Given that the role of Nej1 during NHEJ is unknown, it is difficult to speculate about the nature of such a function. In any event, NHEJ-mediated T–TFs had different molecular requirements than NHEJ at other parts of the genome. In addition, we showed that the fusions formed in a ter1-4L strain did in fact contain a variable number of mutant and/or wild-type telomeric repeats. As observed with DNA blots, the size of telomere fusions in the ter1-4L mutant strains appeared to vary between individual isolates. This variation could contribute to the observed difference in mutant telomere repeat number between the assayed early and late streaks. However, it is possible that loss of one or several telomeric repeats occurred as a result of complications in cloning repetitive DNA sequences in E. coli. It is therefore difficult to draw extensive conclusions from the telomeric sequence data or to speculate about the total number of telomeric repeats present before cloning. Nevertheless, the sequencing data provide a verification of the presence of mutant telomeric repeats, which suggests that a reduced number of Rap1 molecules were bound to the telomeres prior to fusions. This presumably led to uncapping and a possibility for the NHEJ pathway to fuse telomere termini. The PCR-based assay revealed that T–TFs begin to arise immediately, while remaining undetectable on DNA blots. To explain this difference, we favor a model in which T–TFs initially occur both intra- and interchromosomally. In the subsequent streaks, the interchromosomal fusions are selected against due to their disomic nature. Intrachromosomal fusions, which are predicted to be more stable, gradually accumulate. When a majority of cells have formed intrachromosomal fusions, these fusions are possible to detect as discrete bands on DNA blots.
Other studies have shown that S. pombe strains containing T–TFs displayed severe defects in their ability to undergo meiosis (Naito et al. 1998). This defect was interpreted as either a problem inherent to circular chromomsomes or a consequence of a complete lack of telomeric repeats. Our results showed that ter1-AccSna cells containing T–TFs also have severe problems going through meiosis, displaying 100-fold reduced sporulation efficiency. As the T–TFs observed in this study retained telomeric repeats, this result suggests that the circular chromosomes themselves caused the observed meiotic defect. A possible explanation for the rare spores that did result from meiosis is that an even number of crossover events took place between each homologous pair. This would prevent formation of dicentric chromosomes and enable chromosome disjunctions with fused telomeres, thus allowing the cells to proceed through meiosis.
This study provides further evidence for the complex interplay between DSB repair pathways and telomeres. It will be especially interesting to continue exploring the mechanism underlying T–TF formation in K. lactis, as the mechanism appears different from NHEJ at internal DSBs.
Acknowledgments
We thank Mohammed Rahman and Laura Harris for their technical assistance and Sidney Kushner, Mattias Mannervik, and Christos Samakovlis for a critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (GM61645-01) to M.J.M. and by grants from the Swedish Research Council (621-2004-1924) and the Swedish Cancer Society (4592-B03-03XAB) to S.U.Å.
References
- Åström, S. U., and J. Rine, 1998. Theme and variation among silencing proteins in Saccharomyces cerevisiae and Kluyveromyces lactis. Genetics 148: 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åström, S. U., S. M. Okamura and J. Rine, 1999. Yeast cell-type regulation of DNA repair. Nature 397: 310. [DOI] [PubMed] [Google Scholar]
- Baumann, P., E. Podell and T. R. Cech, 2002. Human pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22: 8079–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A. A., and V. Lundblad, 2003. a The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol. Cell. Biol. 23: 8202–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A. A., and V. Lundblad, 2003. b Which end: dissecting Ku's function at telomeres and double-strand breaks. Genes Dev. 17: 2347–2350. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., J. Trueheart, G. Natsoulis and G. R. Fink, 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli, D., A. Smogorzewska, L. Chong and T. de Lange, 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17: 231–235. [DOI] [PubMed] [Google Scholar]
- Callebaut, I., L. Malivert, A. Fischer, J. P. Mornon, P. Revy et al., 2006. Cernunnos interacts with the XRCC4 × DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J. Biol. Chem. 281: 13857–13860. [DOI] [PubMed] [Google Scholar]
- Chan, S. R., and E. H. Blackburn, 2004. Telomeres and telomerase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. W., and E. H. Blackburn, 2003. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell 11: 1379–1387. [DOI] [PubMed] [Google Scholar]
- Chen, X. J., and G. D. Clark-Walker, 1994. sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol. Cell. Biol. 14: 4501–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, M. N., J. H. Wright, A. J. Wolf and V. A. Zakian, 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63: 739–750. [DOI] [PubMed] [Google Scholar]
- Cooper, J. P., E. R. Nimmo, R. C. Allshire and T. R. Cech, 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Dionne, I., and R. J. Wellinger, 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93: 13902–13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois, M. L., Z. W. Haimberger, M. W. McIntosh and D. E. Gottschling, 2002. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone, C., and S. P. Jackson, 1999. Ku, a DNA repair protein with multiple cellular functions? Mutat. Res. 434: 3–15. [DOI] [PubMed] [Google Scholar]
- Feldmann, E., V. Schmiemann, W. Goedecke, S. Reichenberger and P. Pfeiffer, 2000. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28: 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M. G., and J. P. Cooper, 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7: 55–63. [DOI] [PubMed] [Google Scholar]
- Ferreira, M. G., and J. P. Cooper, 2004. Two modes of DNA double-strand break repair are reciprocally regulated through the fission yeast cell cycle. Genes Dev. 18: 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, T. S., and V. A. Zakian, 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Rep. 4: 1215–1226. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15: 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin, N., C. Damon and M. Charbonneau, 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20: 8397–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel, S., M. Larrivee, P. Labrecque and R. J. Wellinger, 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280: 741–744. [DOI] [PubMed] [Google Scholar]
- Herrmann, G., T. Lindahl and P. Schär, 1998. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17: 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H. L., D. Gilley, S. A. Galande, M. P. Hande, B. Allen et al., 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14: 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, G., and J. E. Haber, 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22: 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, S., A. D. Chadha and M. J. McEachern, 2005. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell. Biol. 25: 8064–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel, A., J. O. Sjöstrand and S. U. Åström, 2001. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr. Biol. 11: 1611–1617. [DOI] [PubMed] [Google Scholar]
- Kegel, A., P. Martinez, S. D. Carter and S. U. Åström, 2006. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 34: 1633–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai, K. M., and K. Muniyappa, 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells 2: 443–455. [DOI] [PubMed] [Google Scholar]
- Krauskopf, A., and E. H. Blackburn, 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383: 354–357. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., F. Paques, J. Sylvan and J. E. Haber, 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9: 767–770. [DOI] [PubMed] [Google Scholar]
- Lewis, L. K., F. Storici, S. Van Komen, S. Calero, P. Sung et al., 2004. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166: 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti, G., and E. J. Louis, 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 11: 1373–1378. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and E. H. Blackburn, 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and J. W. Szostak, 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Maringele, L., and D. Lydall, 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16: 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M. J., and E. H. Blackburn, 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376: 403–409. [DOI] [PubMed] [Google Scholar]
- McEachern, M. J., and E. H. Blackburn, 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10: 1822–1834. [DOI] [PubMed] [Google Scholar]
- McEachern, M. J., and S. Iyer, 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7: 695–704. [DOI] [PubMed] [Google Scholar]
- McEachern, M. J., S. Iyer, T. B. Fulton and E. H. Blackburn, 2000. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc. Natl. Acad. Sci. USA 97: 11409–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski, P. A., J. O. Mieczkowska, M. Dominska and T. D. Petes, 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. USA 100: 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, G. T., S. Jin, K. B. Shannon and D. T. Weaver, 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, T., M. Sadaie, J. Kanoh and F. Ishikawa, 2003. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J. Biol. Chem. 278: 1924–1931. [DOI] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H. J., 1938. The remaking of chromosomes. The Collecting Net 8: 182–195. [Google Scholar]
- Naito, T., A. Matsuura and F. Ishikawa, 1998. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 20: 203–206. [DOI] [PubMed] [Google Scholar]
- Nakamura, T. M., J. P. Cooper and T. R. Cech, 1998. Two modes of survival of fission yeast without telomerase. Science 282: 493–496. [DOI] [PubMed] [Google Scholar]
- Natarajan, S., K. Nickles and M. J. McEachern, 2006. Screening for telomeric recombination in wild-type Kluyveromyces lactis. FEMS Yeast Res. 6: 442–448. [DOI] [PubMed] [Google Scholar]
- Nickles, K., and M. J. McEachern, 2004. Characterization of Kluyveromyces lactis subtelomeric sequences including a distal element with strong purine/pyrimidine strand bias. Yeast 21: 813–830. [DOI] [PubMed] [Google Scholar]
- Ooi, S. L., D. D. Shoemaker and J. D. Boeke, 2001. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science 294: 2552–2556. [DOI] [PubMed] [Google Scholar]
- Palladino, F., T. Laroche, E. Gilson, A. Axelrod, L. Pillus et al., 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555. [DOI] [PubMed] [Google Scholar]
- Pardo, B., and S. Marcand, 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24: 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, Z. W. Haimberger et al., 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27: 64–67. [DOI] [PubMed] [Google Scholar]
- Polotnianka, R. M., J. Li and A. J. Lustig, 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8: 831–834. [DOI] [PubMed] [Google Scholar]
- Riha, K., and D. E. Shippen, 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K. B., and T. D. Petes, 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul and M. A. Blasco, 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1: 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schär, P., G. Herrmann, G. Daly and T. Lindahl, 1997. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 11: 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch and T. de Lange, 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644. [DOI] [PubMed] [Google Scholar]
- Stellwagen, A. E., Z. W. Haimberger, J. R. Veatch and D. E. Gottschling, 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17: 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, S. H., and S. P. Jackson, 1997. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 16: 4788–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, S. H., and S. P. Jackson, 2000. Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr. Biol. 10: 165–168. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., A. K. Taggart and V. A. Zakian, 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11: 1328–1335. [DOI] [PubMed] [Google Scholar]
- Tzfati, Y., Z. Knight, J. Roy and E. H. Blackburn, 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 17: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood, D. H., C. Carroll and M. J. McEachern, 2004. Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot. Cell 3: 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus et al., 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414: 666–669. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., A. Smogorzewska and T. de Lange, 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413. [DOI] [PubMed] [Google Scholar]
- Wilson, T. E., U. Grawunder and M. R. Lieber, 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498. [DOI] [PubMed] [Google Scholar]
- Wray, L. V., Jr., M. M. Witte, R. C. Dickson and M. I. Riley, 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]