Abstract
In eukaryotic cells, the inactivation of the cyclic nucleotide signal depends on a complex array of cyclic nucleotide phosphodiesterases (PDEs). Although it has been established that multiple PDE isoenzymes with distinct catalytic properties and regulations coexist in the same cell, the physiological significance of this remarkable complexity is poorly understood. To examine the role of a PDE in cAMP signaling in vivo, we have inactivated the type 4 cAMP-specific PDE (PDE4D) gene, a mammalian homologue of the Drosophila dunce. This isoenzyme is involved in feedback regulation of cAMP levels. Mice deficient in PDE4D exhibit delayed growth as well as reduced viability and female fertility. The decrease in fertility of the null female is caused by impaired ovulation and diminished sensitivity of the granulosa cells to gonadotropins. These pleiotropic phenotypes demonstrate that PDE4D plays a critical role in cAMP signaling and that the activity of this isoenzyme is required for the regulation of growth and fertility.
In organisms as diverse as bacteria and mammals, changes in the intracellular concentration of the second messenger cAMP control a wide range of cell functions including entry and exit from the cell cycle, differentiation, metabolism, and gene expression. Similar to that shown for the Ca2+-dependent signaling pathway (1), it has been hypothesized that the intensity and duration of the cAMP signals determine the specificity of cellular responses to different stimuli (2). The pattern of intracellular cAMP accumulation depends on the activity of cyclic nucleotide phosphodiesterases (PDEs). These enzymes are regulated during hormone and neurotransmitter stimulation (3, 4) in concert with other mechanisms that control cAMP synthesis (5) and have profound effects on cAMP levels and cell responses (3, 4, 6, 7).
To date, at least 20 PDE genes and almost 50 distinct PDE proteins have been described in mammalian cells (8). These isoenzymes are characterized by different biochemical and kinetic properties, subcellular localization, and mechanisms of regulation. However, the reason a cell requires the expression of such a large array of PDEs with an overlapping function is poorly understood. It has been speculated that each isoenzyme serves slightly different roles and its expression is necessary for the specificity of the cAMP signaling or for signal compartmentalization. Here we have addressed the question of this apparent redundancy of the cAMP-degrading machinery by studying the effect of inactivating a single PDE gene.
With some rare exceptions, pharmacological inhibition of PDE activity results in an increased signaling through the cAMP-dependent pathway (9), an effect that mimics the stimulation by hormones and neurotransmitters. Although this idea may hold true for short-term pharmacological manipulations, in vivo long-term cell homeostasis and differentiation is more likely impaired by the disruption of the PDE system. If the expression of distinct PDE isoenzymes is indeed required for proper cAMP signaling, the inactivation of one PDE should produce a loss, rather than a gain of function. This view is supported by the phenotype caused by mutations in the Drosophila melanogaster dunce PDE. Flies deficient in this PDE display impairments of the central nervous system and reproductive functions (10). In the same vein, natural mutations inactivating a PDE expressed in the retina cause retinal degeneration (11).
Although only one dunce PDE has been described in the fly, four orthologous genes (PDE4A, PDE4B, PDE4C, and PDE4D) are present in mice (12), rats (13), and humans (14), and the encoded proteins share considerable homology in their catalytic and regulatory domains (4, 8, 15). The evolutionary significance for this gene duplication is unknown. In vitro and in vivo studies have demonstrated that one of type 4 cAMP-specific PDEs (PDE4D) is activated either through protein kinase A-dependent phosphorylation (16) or through regulation of transcription (17). This dual regulation in endocrine, inflammatory, and neuronal cells determines the intensity and the duration of the cAMP stimulus and promotes adaptive changes such as desensitization (4), at least in vitro.
To dissect the function of different PDEs in vivo, a PDE4 gene widely expressed in different organs of the mouse was inactivated by homologous recombination. The pleiotropic phenotypes produced demonstrate that inactivation of the PDE4D gene causes major loss of cell functions. Thus, we demonstrate that a PDE4D plays a nonredundant role in cyclic nucleotide signaling in vivo.
Materials and Methods
Construction of PDE4D Targeting Vector.
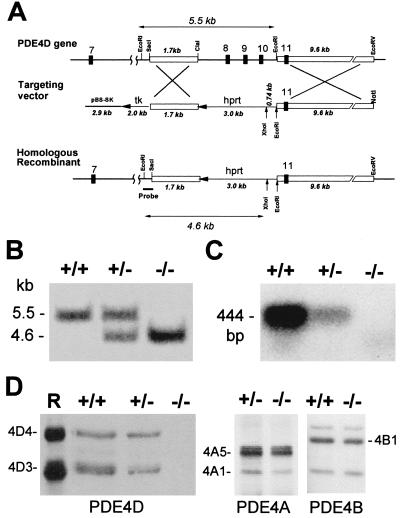
Three overlapping clones corresponding to exons 6–11 of the PDE4D gene were isolated by screening a 129svj lambda FIXII mouse genomic library (Stratagene). A 1.7-kb SacI–ClaI fragment 5′ to exon 8 and a 9.6-kb EcoRI–EcoRV fragment 3′ to exon 10 were isolated and subcloned into a knockout plasmid to generate targeting vectors (Fig. 1A). The knockout plasmid, a gift of D. Repaske, University of Cincinnati, uses pBluescript II SK as a backbone and contains a PGK-hprt cassette (for positive selection) and a pMC1-tk cassette (for negative selection) subcloned at HindIII and KpnI sites, respectively. The targeting vector was constructed by blunt-end ligation of the two genomic fragments at BamHI and ClaI sites of the knockout plasmid so that the hprt and tk genes were in the opposite orientation to the PDE4D genomic sequence (targeting vector I, Fig. 1A). An additional targeting vector also was constructed with the hprt and tk genes in the same orientation as the PDE4D gene (targeting vector II, not shown). In the constructs thus created, the hprt gene substituted for a 3.2-kb genomic fragment that contains exons 8–10 of the gene. The constructs were confirmed by restriction mapping, and the ligation sites were directly validated by DNA sequencing.
Figure 1.
Targeted disruption of the mouse PDE4D locus. (A) Strategy used for targeting the PDE4D gene. The structure of the wild-type PDE4D gene in the region containing exons 7–11 (Top), the targeting vector (Middle), and the homologous recombinant (Bottom) are shown. The targeting vector derived from pBluescript SK+ (pBS-SK) contains 5′ and 3′ flanking regions of homology (open boxes) and is designed to replace the ClaI–EcoRI fragment that contains exons 8–10 with a PGK-hprt cassette. Both hprt and tk genes are in the opposite transcriptional orientation as PDE4D. The probe used for Southern blot analysis of EcoRI/XhoI-digested genomic DNA is indicated. The expected sizes of EcoRI–EcoRI (5.5 kb) and EcoRI–XhoI (4.6 kb) fragments are reported as double arrows. (B) Southern blot analysis of genomic DNA isolated from PDE4D+/+, PDE4D+/−, and PDE4D−/− mouse tail tips, digested with EcoRI and XhoI and hybridized to the probe shown in A. (C) Expression of PDE4D mRNA in mouse brain. Reverse transcription–PCR for poly(A)+ RNA extracted from pooled cortex and cerebellum of PDE4D+/+, PDE4D+/−, and PDE4D−/− mice was carried out as described in Materials and Methods. The 5′ and 3′ primers used in the PCR are specific to the exon 8 and exon 10 sequence of PDE4D, respectively. The 444-bp PCR fragment was detected by Southern blot analysis using an exon 9-specific oligonucleotide probe. (D) Expression of PDE4D, PDE4A, and PDE4B proteins in mouse brain. Cortexes and cerebella dissected from PDE4D+/+, PDE4D+/−, and PDE4D−/− mice were homogenized and immunoprecipitated as described in Materials and Methods. The PDE4D (4D3 and 4D4), PDE4A (4A1 and 4A5), and PDE4B1 proteins were detected by Western blot analysis as described in Materials and Methods. A mixture of recombinant PDE4D3 and PDE4D4 (R) was loaded for control.
Production of PDE4D-Deficient Mice.
The targeting vectors were linearized with NotI and electroporated into E14TG2a embryonic stem (ES) cells (18). Homologous recombinant ES clones were screened by Southern blot analysis. ES cells from positive clones were microinjected into C57BL/6 blastocysts and implanted into pseudopregnant females. The resulting chimeras were mated to C57BL/6 mice to produce F1 progeny with heterozygous genotype (PDE4D+/−). Further crosses between the PDE4D+/− mice yielded F2 generation homozygous mutants (PDE4D−/−) with a mixed 129/Ola and C57BL/6 background. Genotyping of animals was performed on DNA from tail tips by Southern blot analysis. Of 10 chimeras generated from the two PDE4D targeting vectors, three transmitted the disrupted allele to the offspring. Here, we report the data obtained from the mice that were generated from two chimeras of targeting vector I.
Southern Blot Analysis.
DNA prepared from embryonic stem cells and tail tips was digested with XhoI and EcoRI, fractionated by agarose gel electrophoresis, transferred to a nylon membrane (ICN), and then hybridized with the 0.22-kb PCR-generated probe that flanked the 5′ homology region (Fig. 1A). Expected sizes for wild-type and mutant PDE4D are 5.5 kb and 4.6 kb, respectively.
Reverse Transcription–PCR and Southern Blot Analysis of mRNA.
First-strand cDNA was synthesized from 400 ng of poly(A)+ RNA isolated from adult mouse cortex and cerebellum, followed by PCR amplification. PCR primers used were 5′-CGGAACTTGCCTTGATGT-3′ and 5′-ACAGAGGCGTTGTGCTTG-3′ located in exons 8 and 10, respectively. The PCR product was analyzed by Southern blot as described above, except that the membrane was hybridized with an exon 9-specific oligonucleotide probe (5′-CACATGAATCTGCTGGC-3′).
Western Blot Analysis.
Cortex and cerebellum were dissected from adult mouse brain and immediately homogenized as described (19). After centrifugation at 16,000 × g for 20 min, the supernatant was immunoprecipitated with a PDE4D-specific mAb, M3S1, a PDE4A-specific polyclonal antibody, AC55, or a PDE4B-specific polyclonal antibody, K118 (19). The immunoprecipitated PDE4D, PDE4A, and PDE4B proteins were further detected by Western blot analysis using a PDE4D-specific mAb, F34–8F4, AC55, and K118, respectively. The immunoprecipitation and Western blotting procedures were carried out as described (19).
PDE Assay.
Pituitaries, ovaries, and cerebella were dissected from animals and were homogenized as described above. Aliquots of the homogenates were assayed for total PDE activity as well as rolipram-insensitive PDE activity by incubation in the absence or presence of 10 μM rolipram. The PDE assay was performed according to the method of Thompson and Appleman (20) as detailed previously (21). The rolipram-sensitive rolipram-insensitive activity (PDE4 activity) was obtained by subtracting the RI activity from the total activity. Protein concentration was determined by using the Bradford method (22).
Test of Fertility and Ovulation.
Immature (20–35 days of age) or adult cycling females were injected s.c. with 5 units of pregnant mare serum gonadotropin (PMSG, Calbiochem). After 48 h, mice received 5 units i.p. of human chorionic gonadotropin (hCG, Goldline Labs, Fort Lauderdale, FL) to induce ovulation. After 24 h, the ovulated oocytes were retrieved from the ampulla of the oviduct and counted. The ovaries of these mice were fixed with 4% paraformaldehyde. Eight to 10-micron sections were prepared from OCT-embedded ovaries and stained with hematoxylin and eosin according to standard procedures.
cAMP Response to hCG in Granulosa Cells in Vitro.
The cAMP response of granulosa cells to hCG was measured according to the procedure described previously (21). Briefly, adult mice were treated with PMSG for 44 h, and the granulosa cells were collected by puncturing the large antral follicles under a stereomicroscope. After centrifugation, the cells were placed in 1 ml of Hepes-buffered McCoy’s 5A (GIBCO/BRL) culture medium in the absence or presence of hCG (100 ng/ml, National Institute of Diabetes and Digestive and Kidney Diseases), follicle-stimulating hormone (250 ng/ml, National Institute of Diabetes and Digestive and Kidney Diseases), or forskolin (100 μM). To terminate the incubation, cultured cells were centrifuged at 1,000 × g for 10 min, and 0.1% trichloroacetic acid in 95% ethanol was added to the cell pellet. After 30-min extraction, the supernatant was recovered by centrifugation at 3,000 × g for 10 min and dried under vacuum. The cAMP levels were determined by RIA (23).
Results
Targeted Disruption of the PDE4D Gene.
Of the four PDE4 loci present in the mouse, we report here the inactivation of the PDE4D gene. The PDE4D locus is composed of at least 18 exons distributed over 150 kb of genomic DNA. Multiple transcriptional units and alternate splicing generate five transcripts that encode five distinct proteins with divergent amino termini (8). Because of the presence of multiple promoters and to ensure an inactivation of all PDE4D variants, exons encoding the catalytic domain were chosen as the target for recombination (Fig. 1A). Exons 8–10 of the gene contain sequences highly conserved among PDEs, and point mutations or deletions in the region produce catalytically inactive PDE4D proteins (24).
Genotyping of the F2 generation mice demonstrated the predicted shift in the size of a restriction fragment from 5.5 to 4.6 kb (Fig. 1B). The PDE4D mRNA levels were measured by reverse transcription–PCR followed by Southern blot analysis. Although a PDE4D mRNA was readily detected in wild-type (PDE4D+/+) mice, a significant decrease of mRNA and no mRNA was observed in the PDE4D+/− and PDE4D−/− mice, respectively (Fig. 1C). The loss of PDE4D mRNA was reflected in the absence of PDE4D protein expression. In the brain, where PDE4D is expressed at high levels, none of the PDE4D variants could be detected in the PDE4D−/− mice after immunoprecipitation and Western blot analysis using antibodies that recognize epitopes at the carboxyl terminus (Fig. 1D) and at the amino terminus of PDE4D (data not shown). Conversely, the products of the PDE4A and PDE4B genes were expressed at a comparable level in the PDE4D+/+ and PDE4D−/− mice (Fig. 1D).
Effects of the Disrupted PDE4D Gene on PDE Activity.
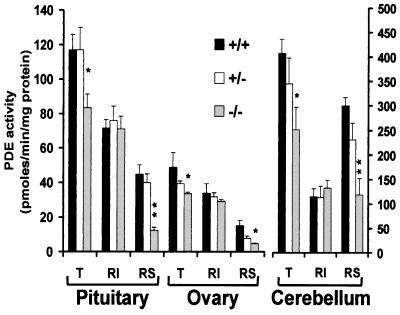
To determine whether the inactivation of the PDE4D gene is associated with a decrease in PDE activity, cAMP hydrolysis was measured in extracts of cerebella, pituitaries, and ovaries in the absence or presence of rolipram, a PDE4-specific inhibitor (Fig. 2). Total PDE activity was decreased in the PDE4D−/− mice in all regions examined. This decrease was entirely the result of a decrease in rolipram-sensitive activity, and no significant changes could be observed for the rolipram-insensitive activity (Fig. 2). Although activity decreased to different extents depending on the organ, similar findings were obtained with cerebral cortex, hippocampus, liver, lung, and spleen (data not shown). It also was observed that in some tissues, such as pituitary and cerebellum, only a minimal decrease in PDE activity was detected in the heterozygous mice. Because we have shown that PDE4D expression is regulated by cAMP (25), inactivation of one PDE4D allele may cause a cAMP-dependent compensatory increase in transcription from the wild-type allele.
Figure 2.
PDE activity in different mouse organs. Pituitaries, ovaries, and cerebella were dissected from PDE4D+/+, PDE4D+/−, and PDE4D−/− mice and were homogenized as described in Materials and Methods. Aliquots of the homogenates were assayed for total PDE activity (T) or rolipram-insensitive (RI) PDE activity in the absence or presence of 10 μM rolipram, respectively. The rolipram-sensitive (RS) activity was obtained by subtracting the RI value from the T value. Data are the mean ± SEM (n ≥ 3 mice/genotype). Significant differences in the total activity and in the RS activity between PDE4D+/+ and PDE4D−/− mice were determined by t test and are indicated by single (*, P < 0.05) and double asterisks (**, P < 0.005).
Viability of the PDE4D−/− Mice.
Genotyping of F2 mice at 30 days of age (D30) demonstrated a distortion of the Mendelian inheritance of the null allele, with only 13.8%, rather than the expected 25%, of PDE4D−/− mice recovered (Table 1). A similar result also was obtained for the mice derived from targeting vector II, ruling out the possibility that phenotype is caused by spurious rearrangements at the site of recombination. Further genotyping of D0 (newborn), D7, and D14 pups demonstrated that most PDE4D−/− mice died between D0 and D14 (Table 1). This phenotype was exacerbated when the PDE4D null allele was transferred to an 129svj background (9.5% homozygous null recovered at D30). Thus, more than 40% of the PDE4D−/− mice died within 4 weeks after birth.
Table 1.
Transmission of the PDE4D null allele to the F2 generation
| Age | Mice genotyped | +/+
|
+/−
|
−/−
|
|||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| D0 | 93 | 24 | 25.8 | 40 | 43 | 29 | 31.2 |
| D7 | 69 | 19 | 27.5 | 33 | 47.8 | 17 | 24.6 |
| D14 | 65 | 17 | 26.2 | 38 | 58.5 | 10 | 15.4 |
| D30 | 347 | 94 | 27.1 | 205 | 59.1 | 48 | 13.8* |
Mice were produced by mating F1 heterozygous males and females.
P < 0.001 by χ2-test, with the expected genotype ratio of 25% of +/+, 50% of +/−, and 25% of −/−.
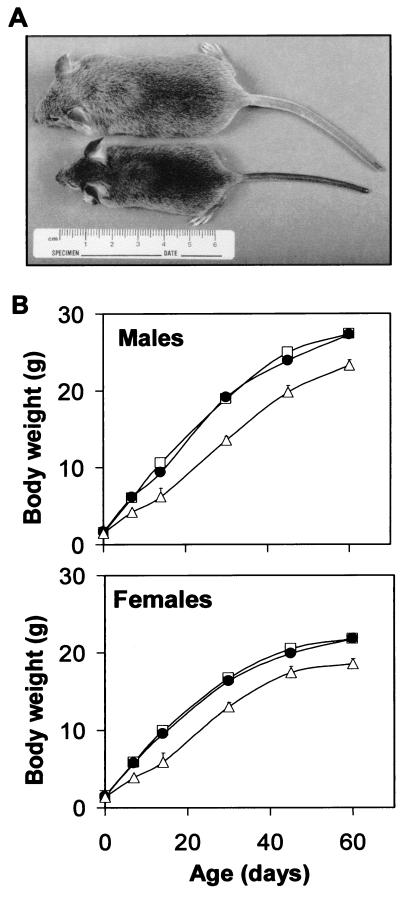
Growth Retardation of the PDE4D−/− Pups.
Surviving PDE4D−/− pups had retarded growth, exhibiting a 30–40% decrease in body weight at 1–2 weeks after birth (Fig. 3). Growth rate returned to normal after 2 weeks, but the weight of the adult mice remained lower than normal (Fig. 3B). This phenotype again was confirmed in the mice that were derived from targeting vector II (data not shown). The decrease in body weight was caused by a proportional decrease in muscle and bone mass and internal organ weight with the exception of cortex and cerebellum (data not shown). These two parts of the brain had weights comparable to those of wild-type littermates. The PDE4D+/− mice had a phenotype similar to the PDE4D+/+ mice, essentially excluding the possibility that truncated PDE4D produced from the disrupted allele exerts dominant negative effects. The decrease in weight of the PDE4D−/− mice was associated with a decrease in circulating insulin-like growth factor I (IGF-I) levels (ng/ml; PDE4D+/− mice: 372 ± 35, n = 15; PDE4D−/− mice: 213 ± 45, n = 6; P < 0.05; measured at D15), suggesting that the GH-IGF-I axis is impaired in immature mice deficient in PDE4D.
Figure 3.
Growth of PDE4D-deficient mice. (A) Comparison of a PDE4D−/− mouse (Lower) and a control littermate (Upper) at D34. (B) Growth curves of PDE4D+/+ (■), PDE4D+/− (●), and PDE4D−/− (▵) mice. Each point represents the mean ± SEM (n = 3–99 mice/genotype).
Effects of the Disrupted PDE4D Gene on Female Fertility.
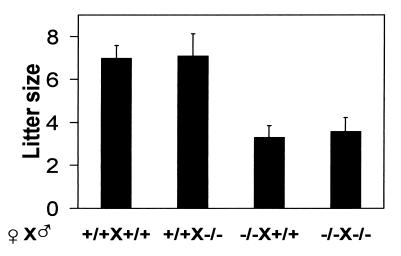
Mating of PDE4D−/− females and males produced smaller litter sizes than the control PDE4D+/+ littermates. To further determine whether, in addition to decreased viability, reduced fertility of the homozygous pairs contributes to the small litter size, PDE4D−/− females and males were mated to PDE4D+/+ mice and the litter size was determined on D0 (Fig. 4). Because the pups generated by these matings are all heterozygous, the litter size should no longer be affected by the decreased viability of the PDE4D−/− litters. Although PDE4D−/− males produced a similar number of pups to PDE4D+/+ males, PDE4D−/− females produced significantly fewer pups, demonstrating that a reduced fertility of the female is a major cause for the small litter size from homozygous mating.
Figure 4.
Female fertility of PDE4D-deficient mice. The litter size from each mating pair was determined at the day of birth. The number of mating pairs in each group is from left to right 11, 9, 13, and 17. Data are the mean ± SEM. P < 0.001, PDE4D−/− females versus wild-type females determined by ANOVA.
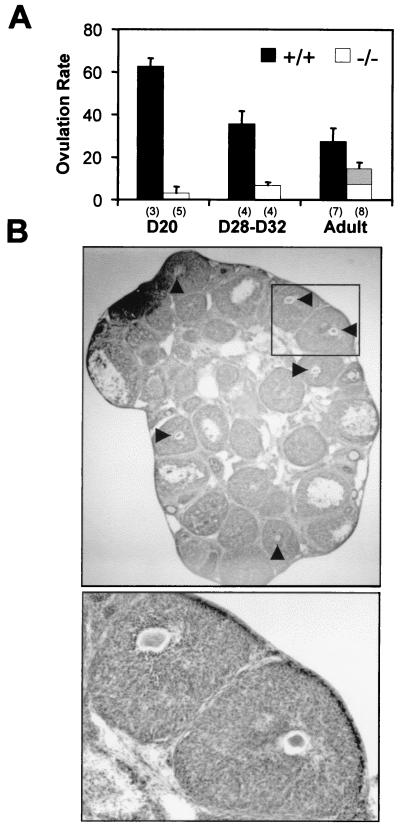
Using in situ hybridization in the ovary, we have shown that PDE4D mRNA is expressed at high levels in the mural granulosa cells, but not in the cumulus or theca cells or in the oocytes (26). Furthermore, follicle-stimulating hormone and luteinizing hormone regulate PDE4 activity in vitro (21), and in vivo studies have demonstrated that PDE4 activity changes in synchrony with the follicular cycle (27). These findings suggest that an impaired follicular function is the cause of the reduced fertility in PDE4D−/− females. This possibility was investigated by measuring the ovulation rate in PDE4D−/− females after superovulation induction. In contrast to the wild-type pubertal littermates, the PDE4D−/− females (D20 or D28–32) yielded few oocytes in response to PMSG-hCG stimulation (Fig. 5A). The D28–32 null mice have a greater weight than the D20 wild-type mice, ruling out the possibility that the ovulation failure is a consequence of the immaturity of the mice. Although the ovulation rate of adult cycling PDE4D−/− females was increased in comparison to pubertal mice (Fig. 5A), more than 50% of the oocytes recovered from the ampulla of the oviducts showed clear signs of degeneration with disrupted metaphase or oocyte fragmentation (data not shown). In contrast, degeneration was only occasionally observed in the wild-type females.
Figure 5.
Ovulation of PDE4D-deficient mice. (A) Ovulation rate (oocytes/mouse) of mice at different ages of development. Wild-type (+/+) and homozygous null (−/−) mice were treated with PMSG and hCG as described in Materials and Methods. Ovulated oocytes were recovered from the ampulla of the oviducts. Data are the mean ± SEM. PDE4D−/− mice are significantly different from PDE4D+/+ mice with P < 0.001 (analyzed by ANOVA). The gray area within the bar represents the degenerated oocytes recovered in the adult mice. Numbers in the brackets indicate the number of mice used in each group. (B) Histological appearance of ovaries from pubertal PDE4D−/− mice (Upper, ×16; Lower, ×52). Mice were treated for superovulation, and ovaries were dissected, fixed, sectioned, and stained. Luteinized follicles with trapped oocytes are indicated by arrowheads.
Histological examination of the ovaries from the D20 superovulated mice demonstrated that there was a major increase in the number of follicles with oocytes entrapped (Fig. 5B), confirming a defect in ovulation. Some of these follicles have undergone luteinization on the basis of their morphological appearance (Fig. 5B). Conversely, the ovaries from adult superovulated PDE4D−/− mice were grossly normal, with all stages of follicular maturation present.
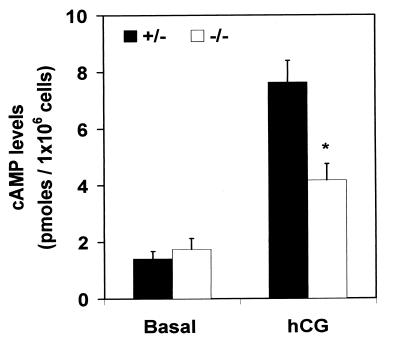
When oocytes of adult PDE4D−/− mice were removed from the antral follicles and cultured in vitro, no major defect was observed and all oocytes resumed meiosis and were arrested in metaphase II (data not shown). This arrest was maintained for at least 24 h, indicating that the degeneration of the oocyte observed in vivo is not caused by a deficit in mos (28) or M-phase promoting factor/cytostatic factor, both required for maintaining the meiotic arrest (29). This finding, together with the observation that PDE4D is not expressed in the oocytes, indicates that the degeneration is not caused by a primary defect in the oocyte but rather by an altered granulosa cell function in the PDE4D−/− mice. To test this possibility, granulosa cells isolated from PMSG-treated PDE4D+/− and PDE4D−/− mice were cultured in the absence or presence of hCG, and the intracellular cAMP accumulation was measured. Granulosa cells derived from the PDE4D−/− mice displayed a moderate increase in basal cAMP levels (P = 0.08, Fig. 6). Surprisingly, in spite of a decrease in the rolipram-sensitive PDE activity in granulosa cells from the null mice (pmol/min per mg protein, PDE4D+/−: 4.74 ± 0.25; PDE4D−/−: 3.02 ± 0.39, n = 3; P < 0.05), a significant reduction in response to hCG was observed with a 50% decrease in cAMP accumulation detected after 1 h (Fig. 6). A similar decrease in cAMP accumulation was observed when follicle-stimulating hormone was used to stimulate the PDE4D−/− granulosa cells, whereas the response to forskolin was not affected (data not shown). This latter finding indicates that the machinery involved in the cAMP synthesis is not affected in the granulosa cells of the null mice. A detailed time course study of hCG stimulation showed no major changes in the temporal pattern of cAMP accumulation (data not shown). An impaired granulosa cell response to gonadotropins was further indicated by the finding that uterine weight after PMSG treatment in the adult PDE4D−/− mice is consistently lower than that of heterozygous mice (PDE4D+/− mice: 98.56 ± 2.57 mg, n = 20; PDE4D−/− mice: 74.28 ± 6.24 mg, n = 8, P = 0.005).
Figure 6.
cAMP response to hCG stimulation in granulosa cells. Granulosa cells isolated from antral follicles of the PMSG-treated mice were cultured in the absence (basal) or presence of 100 ng/ml hCG. After 1-h incubation, the intracellular cAMP accumulation was measured by RIA. Data are the mean ± SEM (n = 4 independent experiments each carried out in triplicate). The difference between PDE4D+/− and PDE4D−/− mice in response to hCG is significant (P < 0.001 determined by paired t test, *).
Discussion
With the targeted disruption of the PDE4D gene, we have demonstrated that PDE4D plays a unique and nonredundant role in the cell. Loss of its function cannot be compensated for by any of the cognate PDE4s or members of other PDE families expressed. Thus, inactivation of PDE4D has a wide array of pleiotropic phenotypes, including growth retardation and reduction in viability and female fertility, demonstrating a critical and indispensable role of this enzyme and its regulations in cell homeostasis. Moreover, inactivation of a single PDE4 gene results in altered cAMP signaling and loss of functions in the cell.
Female mice deficient in PDE4D display a reduced fertility with more than a 50% decrease in litter size. All of the data accumulated point to a defect in the granulosa cell function as the underlying cause of the infertility. Gonadotropin responses including cAMP accumulation in granulosa cells, estrogen production measured as uterine weight, and ovulation rate all were decreased in the PDE4D null mice, suggesting that granulosa cell differentiation is disrupted after the inactivation of PDE4D. The consequent reduced number of ovulated oocytes and their degeneration are sufficient to account for the reduced fertility of the adult PDE4D−/− females. Even though the superovulation protocol used in our studies eliminates a possible pituitary contribution to the ovulation phenotype, pituitary contribution to the reduced fertility of the adult cannot be ruled out at this time.
Although inactivation of PDE4D does not cause a complete arrest of follicular development, a significant decrease in the ovulation process was evident at all ages studied. This phenotype is similar to that produced by targeted disruption of other genes expressed in granulosa cells, including the progesterone receptor, estrogen receptor β, transcriptional factor C/EBPβ (CCAAT/enhancer-binding protein β), aromatase, cyclin D2, and cyclooxygenase II (30). A common property of these genes is that they all are induced by gonadotropins via activation of the cAMP signaling pathway, opening the possibility that their expression is altered in the PDE4D null mice where the inactivation of PDE4D disrupts cAMP signaling.
It should be emphasized that in spite of the fact that PDE activity is reduced in granulosa cells of the PDE4D−/− mice, rather than increased, the cAMP response to gonadotropin was significantly reduced. This apparent paradox can be reconciled by hypothesizing that PDE4D regulation in granulosa cells has a protective effect on responsiveness, and that inactivation of this PDE4D negative feedback causes a permanent desensitized state of the gonadotropin signal transduction. For instance, a chronic increase in cAMP resulting from the PDE4D inactivation may produce a condition similar to that produced by the targeted disruption of the RIIβ regulatory subunit of protein kinase A (31). In this latter case, loss of RII subunit also causes a decrease, rather than an increase, in the C subunit activity because of an accelerated degradation. Moreover, an altered duration and intensity of a transient cAMP accumulation may have a major impact in the ovarian follicle, where cAMP is thought to diffuse through gap junctions between granulosa cells themselves and between granulosa cells and oocytes (32). In addition to compromising the normal dynamics of follicular differentiation, an altered half-life and diffusion of cAMP may abrogate differences between mural and cumulus granulosa cell functions that are crucial for oocyte maturation and ovulation (32). The reduced viability of the oocytes that are ovulated in PDE4D−/− adult mice probably is caused by the uncoupling of granulosa cell and oocyte maturation. In view of our finding that PDE4 inhibitors cause oocyte maturation in follicle culture in the absence of gonadotropin stimulation (26), it is possible that resumption of meiosis in the PDE4D null mice is no longer synchronized with final maturation of the follicle.
Our findings are strikingly similar to the phenotype caused by mutations in the Drosophila dunce PDE. The defect in egg deposition in the mutant flies is caused by an impaired function of both the egg and nursing cells (33). Thus, PDE4D plays a critical role in the ovarian follicle, and this function is conserved from Drosophila to mammals.
In endocrine glands, receptor/G protein-activating mutations produce a phenotype of hyperstimulation or end-organ autonomous function (34). A similar phenotype would be predicted by inactivation of a PDE gene because the decrease in the rate of cAMP degradation would be expected to cause increased cAMP signaling and chronic stimulation of the endocrine gland. This, however, was not the case in the PDE4D-deficient mice. These mice present a phenotype of disrupted cAMP signaling in granulosa cells rather than hyperfunction. Similar decreased sensitivity or altered cAMP signaling may explain the reduced growth of the PDE4D−/− mice. We could speculate that an altered cAMP accumulation in somatotrophs of the pituitary disrupts the growth hormone releasing hormone-growth hormone axis as it has been demonstrated for the lit/lit mouse (35). Thus, the phenotype of the PDE4D null mice opens the possibility that endocrine disorders characterized by end-organ resistance are caused by inherited recessive mutations that disrupt the PDE4D function.
Measurements of the PDE activity in different organs from PDE4D−/− mice demonstrate a marked decrease in the rolipram-sensitive activity. Together with the observed phenotypes, these findings demonstrate that other PDE4s expressed in mammals cannot compensate for the loss of PDE4D and that the function of this gene does not completely overlap with the function of other PDE4s. Differences in expression, regulation, and localization of the different PDE4s account for these divergent functions (4). This observation has important pharmacological implications. Because PDE4 inhibitors are being developed as anti-inflammatory agents for asthma, chronic obstructive pulmonary disease, and other inflammatory disorders, our data hold promise for specific inhibition of a single PDE4 gene product as a viable strategy for pharmacological intervention (36).
Acknowledgments
We are indebted to Drs. Alex Tsafriri, Aaron J. W. Hsueh, Richard Roth, and Anita Payne for discussion and critical review of the manuscript. We also thank Linda Lan and Gu Zhang for their skillful technical assistance. This work was supported by National Institutes of Health Grants HD20788, HD31544, and U54-HD31398-02. F.J.R. was supported by a fellowship from Natural Sciences and Engineering Research Council (Canada).
Abbreviations
- PDE
cyclic nucleotide phosphodiesterase
- PDE4
type 4 cAMP-specific PDE
- PMSG
pregnant mare serum gonadotropin
- hCG
human chorionic gonadotropin
References
- 1.Dolmetsch R E, Lewis R S, Goodnow C C, Healy J I. Nature (London) 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 2.Hempel C M, Vincent P, Adams S R, Tsien R Y, Selverston A I. Nature (London) 1996;384:166–169. doi: 10.1038/384166a0. [DOI] [PubMed] [Google Scholar]
- 3.Beavo J A. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 4.Conti M, Nemoz G, Sette C, Vicini E. Endocr Rev. 1995;16:370–389. doi: 10.1210/edrv-16-3-370. [DOI] [PubMed] [Google Scholar]
- 5.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 6.Swinnen J V, D’Souza B, Conti M, Ascoli M. J Biol Chem. 1991;266:14383–14389. [PubMed] [Google Scholar]
- 7.Conti M, Toscano M V, Petrelli L, Geremia R, Stefanini M. Endocrinology. 1983;113:1845–1853. doi: 10.1210/endo-113-5-1845. [DOI] [PubMed] [Google Scholar]
- 8.Conti M, Jin S-L C. Prog Nucleic Acid Res Mol Biol. 1999;63:1–38. doi: 10.1016/s0079-6603(08)60718-7. [DOI] [PubMed] [Google Scholar]
- 9.Thompson W J. Pharmacol Ther. 1991;51:13–33. doi: 10.1016/0163-7258(91)90039-o. [DOI] [PubMed] [Google Scholar]
- 10.Dudai Y, Jan Y N, Byers D, Quinn W G, Benzer S. Proc Natl Acad Sci USA. 1976;73:1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowes C, Li T, Danciger M, Baxter L C, Applebury M L, Farber D B. Nature (London) 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- 12.Cherry J A, Davis R L. J Neurobiol. 1995;28:102–113. doi: 10.1002/neu.480280109. [DOI] [PubMed] [Google Scholar]
- 13.Swinnen J V, Joseph D R, Conti M. Proc Natl Acad Sci USA. 1989;86:5325–5329. doi: 10.1073/pnas.86.14.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger G, Michaeli T, Martins T, St. John T, Steiner B, Rodgers L, Riggs M, Wigler M, Ferguson K. Mol Cell Biol. 1993;13:6558–6571. doi: 10.1128/mcb.13.10.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houslay M D, Sullivan M, Bolger G B. Adv Pharmacol. 1998;44:225–342. doi: 10.1016/s1054-3589(08)60128-3. [DOI] [PubMed] [Google Scholar]
- 16.Sette C, Conti M. J Biol Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 17.Swinnen J V, Tsikalas K E, Conti M. J Biol Chem. 1991;266:18370–18377. [PubMed] [Google Scholar]
- 18.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 19.Iona S, Cuomo M, Bushnik T, Naro F, Sette C, Hess M, Shelton E R, Conti M. Mol Pharmacol. 1998;53:23–32. doi: 10.1124/mol.53.1.23. [DOI] [PubMed] [Google Scholar]
- 20.Thompson W J, Appleman M M. Biochemistry. 1971;10:311–316. [PubMed] [Google Scholar]
- 21.Conti M, Kasson B G, Hsueh A J W. Endocrinology. 1984;114:2361–2368. doi: 10.1210/endo-114-6-2361. [DOI] [PubMed] [Google Scholar]
- 22.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Harper J F, Brooker G. J Cyclic Nucleotide Res. 1975;1:207–218. [PubMed] [Google Scholar]
- 24.Jin S-L C, Swinnen J V, Conti M. J Biol Chem. 1992;267:18929–18939. [PubMed] [Google Scholar]
- 25.Swinnen J V, Joseph D R, Conti M. Proc Natl Acad Sci USA. 1989;86:8197–8201. doi: 10.1073/pnas.86.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsafriri A, Chun S Y, Zhang R, Hsueh A J W, Conti M. Dev Biol. 1996;178:393–402. doi: 10.1006/dbio.1996.0226. [DOI] [PubMed] [Google Scholar]
- 27.Schmidtke J, Meyer H, Epplen J T. Acta Endocrinol. 1980;95:404–411. doi: 10.1530/acta.0.0950404. [DOI] [PubMed] [Google Scholar]
- 28.Colledge W H, Carlton M B L, Udy G B, Evans M J. Nature (London) 1994;370:65–68. doi: 10.1038/370065a0. [DOI] [PubMed] [Google Scholar]
- 29.Singh B, Arlinghaus R B. Prog Cell Cycle Res. 1997;3:251–259. doi: 10.1007/978-1-4615-5371-7_20. [DOI] [PubMed] [Google Scholar]
- 30.Elvin J A, Matzuk M M. Rev Reprod. 1998;3:183–195. doi: 10.1530/ror.0.0030183. [DOI] [PubMed] [Google Scholar]
- 31.Adams M R, Brandon E P, Chartoff E H, Idzerda R L, Dorsa D M, McKnight G S. Proc Natl Acad Sci USA. 1997;94:12157–12161. doi: 10.1073/pnas.94.22.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eppig J J, Chesnel F, Hirao Y, O’Brien M J, Pendola F L, Watanabe S, Wigglesworth K. Hum Reprod. 1997;12:127–132. [PubMed] [Google Scholar]
- 33.Bellen H J, Gregory B K, Olsson C L, Kiger J A J. Dev Biol. 1987;121:432–444. doi: 10.1016/0012-1606(87)90180-1. [DOI] [PubMed] [Google Scholar]
- 34.Ringel M D, Schwindinger W F, Levine M A. Medicine (Baltimore) 1996;75:171–184. doi: 10.1097/00005792-199607000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Godfrey P, Rahal J O, Beamer W G, Copeland N G, Jenkins N A, Mayo K E. Nat Genet. 1993;4:227–232. doi: 10.1038/ng0793-227. [DOI] [PubMed] [Google Scholar]
- 36.Torphy T J. Am J Respir Crit Care Med. 1998;157:351–370. doi: 10.1164/ajrccm.157.2.9708012. [DOI] [PubMed] [Google Scholar]