Abstract
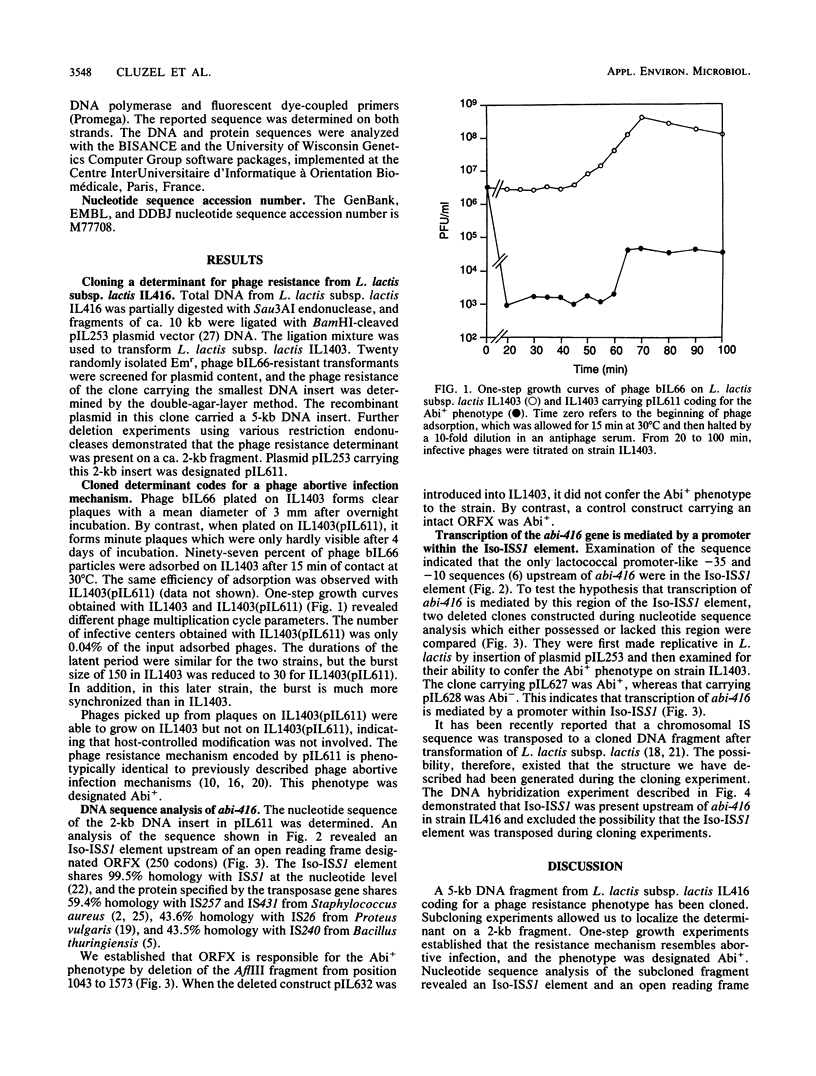
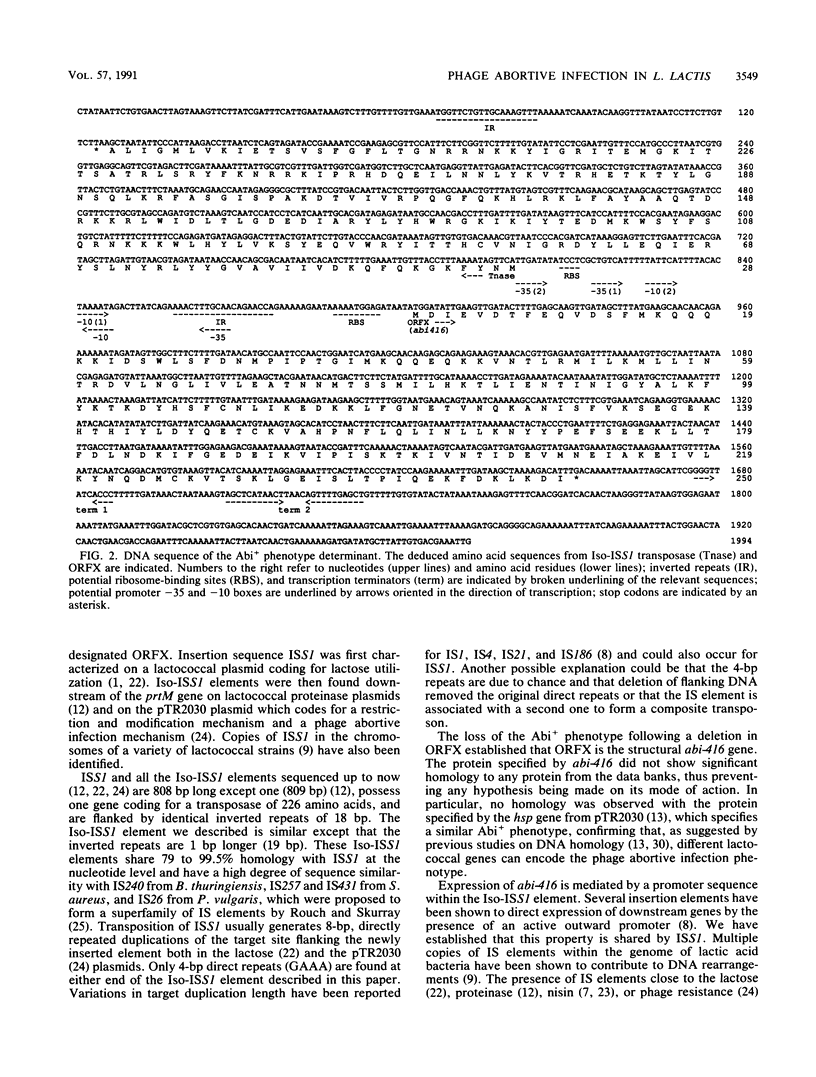
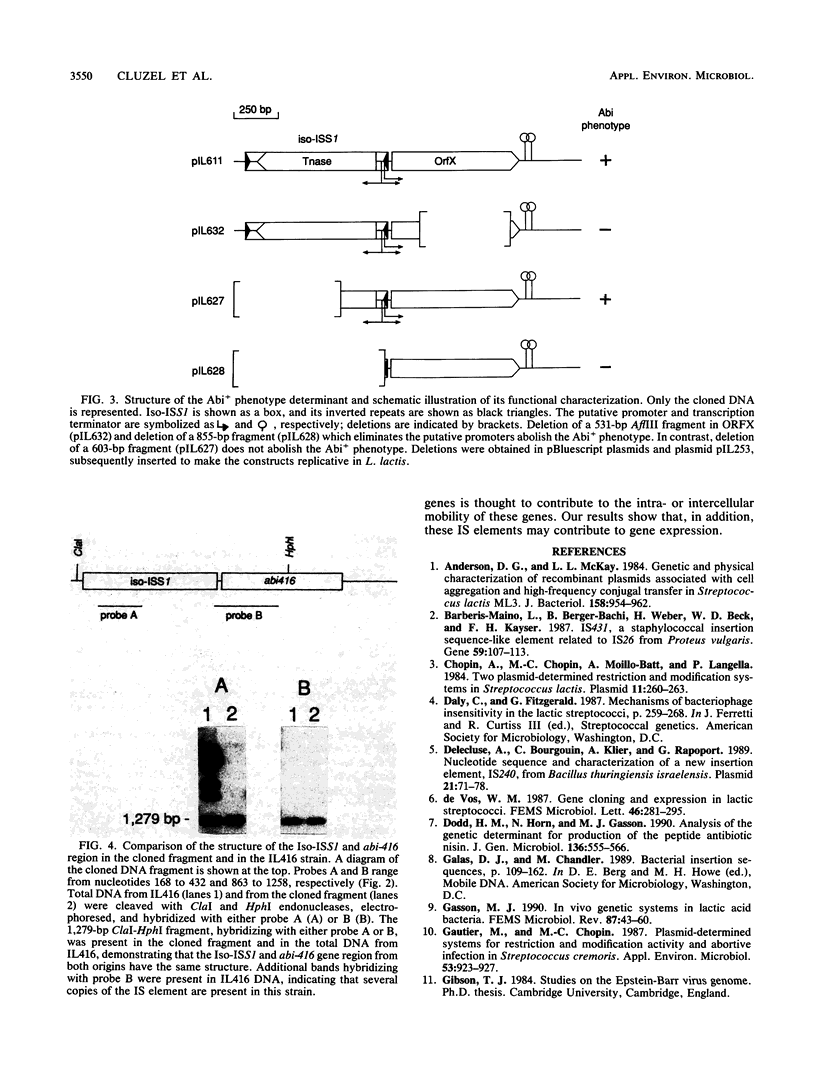
A 5-kb DNA fragment conferring a phage abortive infection phenotype (Abi+) has been cloned from Lactococcus lactis subsp. lactis IL416. The Abi+ determinant was subcloned on a 2-kb fragment which carried an Iso-ISS1 element and an open reading frame of 753 bp designated ORFX. Deletion within ORFX entailed the loss of the Abi+ phenotype, establishing that ORFX is the structural abi-416 gene. The expression of abi-416 was shown to be mediated by the Iso-ISS1 element, which contains a sequence fitting the consensus sequence for gram-positive promoters.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984 Jun;158(3):954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis-Maino L., Berger-Bächi B., Weber H., Beck W. D., Kayser F. H. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene. 1987;59(1):107–113. doi: 10.1016/0378-1119(87)90271-x. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Delecluse A., Bourgouin C., Klier A., Rapoport G. Nucleotide sequence and characterization of a new insertion element, IS240, from Bacillus thuringiensis israelensis. Plasmid. 1989 Jan;21(1):71–78. doi: 10.1016/0147-619x(89)90088-7. [DOI] [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. In vivo genetic systems in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):43–60. doi: 10.1111/j.1574-6968.1990.tb04878.x. [DOI] [PubMed] [Google Scholar]
- Gautier M., Chopin M. C. Plasmid-Determined Systems for Restriction and Modification Activity and Abortive Infection in Streptococcus cremoris. Appl Environ Microbiol. 1987 May;53(5):923–927. doi: 10.1128/aem.53.5.923-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandrikman A. J., van Leeuwen C., Kok J., Vos P., de Vos W. M., Venema G. Insertion elements on lactococcal proteinase plasmids. Appl Environ Microbiol. 1990 Jun;56(6):1890–1896. doi: 10.1128/aem.56.6.1890-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990 Jul;56(7):2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Bohanon M. J., Polzin K. M., Rule P. L., Baldwin K. A. Localization of Separate Genetic Loci for Reduced Sensitivity towards Small Isometric-Headed Bacteriophage sk1 and Prolate-Headed Bacteriophage c2 on pGBK17 from Lactococcus lactis subsp. lactis KR2. Appl Environ Microbiol. 1989 Oct;55(10):2702–2709. doi: 10.1128/aem.55.10.2702-2709.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet B., Iida S., Shepherd J., Arber W. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res. 1983 Sep 24;11(18):6319–6330. doi: 10.1093/nar/11.18.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. C., Steele J. L., Daly C., McKay L. L. Concomitant conjugal transfer of reduced-bacteriophage-sensitivity mechanisms with lactose- and sucrose-fermenting ability in lactic streptococci. Appl Environ Microbiol. 1988 Aug;54(8):1951–1956. doi: 10.1128/aem.54.8.1951-1956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol. 1991 Mar;57(3):734–743. doi: 10.1128/aem.57.3.734-743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987 Dec;169(12):5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch P. J., Beerthuyzen M. M., de Vos W. M. Nucleotide sequence of IS904 from Lactococcus lactis subsp. lactis strain NIZO R5. Nucleic Acids Res. 1990 Jul 25;18(14):4253–4254. doi: 10.1093/nar/18.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. Characterization of insertion sequence IS946, an Iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990 Aug;172(8):4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch D. A., Skurray R. A. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene. 1989;76(2):195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- Sanders M. E. Phage resistance in lactic acid bacteria. Biochimie. 1988 Mar;70(3):411–422. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- Simon D., Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988 Apr;70(4):559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- Simon D., Rouault A., Chopin M. C. High-efficiency transformation of Streptococcus lactis protoplasts by plasmid DNA. Appl Environ Microbiol. 1986 Aug;52(2):394–395. doi: 10.1128/aem.52.2.394-395.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J. L., Murphy M. C., Daly C., McKay L. L. DNA-DNA homology among lactose- and sucrose-fermenting transconjugants from Lactococcus lactis strains exhibiting reduced bacteriophage sensitivity. Appl Environ Microbiol. 1989 Sep;55(9):2410–2413. doi: 10.1128/aem.55.9.2410-2413.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]