Abstract
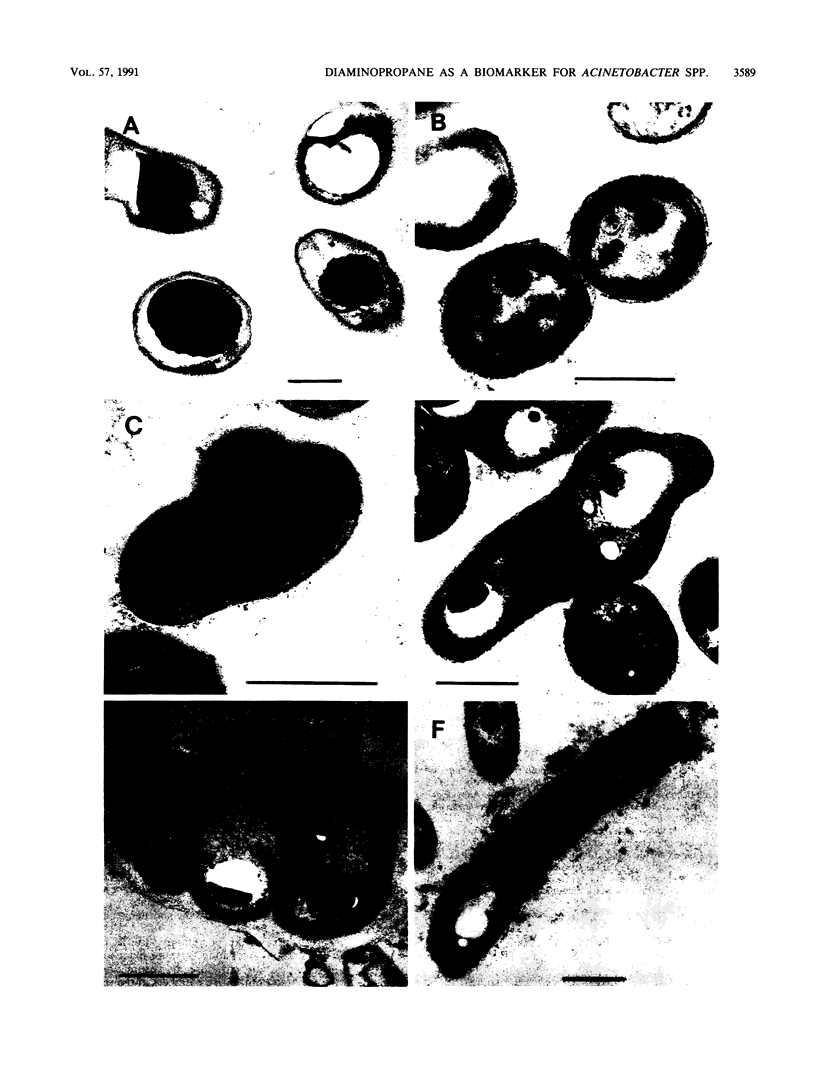
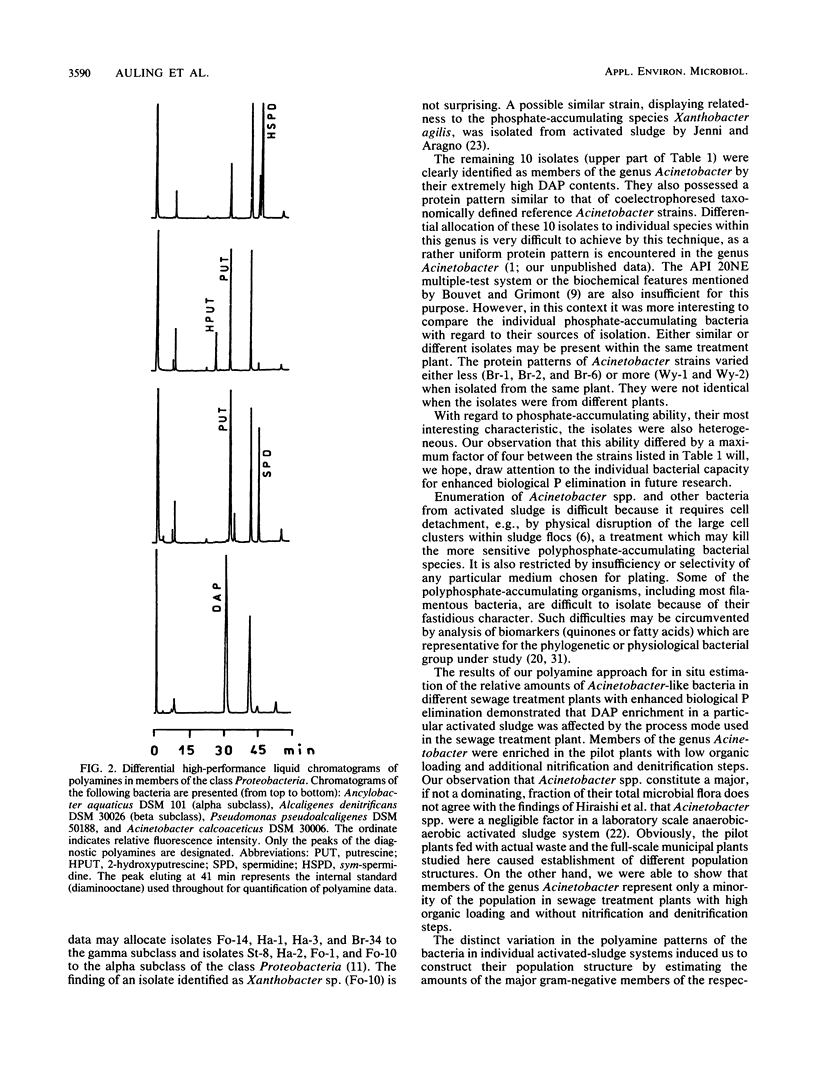
Polyphosphate-accumulating gram-negative bacteria were isolated from different anaerobic-aerobic activated sludge systems with diverse processes for enhanced biological phosphorus (P) elimination. Of 22 isolates, 10 were allocated to the genus Acinetobacter by using multiple-test systems and soluble protein and polyamine patterns. As diaminopropane (DAP) appears to be the characteristic main polyamine compound produced by Acinetobacter spp., it was used as a biomarker for the genus. The high DAP contents of representative samples from municipal wastes with enhanced biological P elimination indicated that Acinetobacter spp. can be dominant organisms in sewage treatment plants with low organic loading and nitrification and denitrification steps. Contrary to accepted opinion, sludge from treatment plants with efficient P removal and high organic loading had a low DAP content, indicating that bacteria other than Acinetobacter spp. are responsible for enhanced biological P elimination in these plants.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander M., Ismail F., Jackman P. J., Noble W. C. Fingerprinting Acinetobacter strains from clinical sources by numerical analysis of electrophoretic protein patterns. J Med Microbiol. 1984 Aug;18(1):55–64. doi: 10.1099/00222615-18-1-55. [DOI] [PubMed] [Google Scholar]
- Baumann P., Doudoroff M., Stanier R. Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J Bacteriol. 1968 May;95(5):1520–1541. doi: 10.1128/jb.95.5.1520-1541.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse H., El-Banna T., Auling G. Evaluation of different approaches for identification of xenobiotic- degrading pseudomonads. Appl Environ Microbiol. 1989 Jun;55(6):1578–1583. doi: 10.1128/aem.55.6.1578-1583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERLACH E., DEUTICKE B. EINE EINFACHE METHODE ZUR MIKROBESTIMMUNG VON PHOSPHAT IN DER PAPIERCHROMATOGRAPHIE. Biochem Z. 1963 Jul 26;337:477–479. [PubMed] [Google Scholar]
- Golecki J. R., Schumacher A., Drews G. The differentiation of the photosynthetic apparatus and the intracytoplasmic membrane in cells of Rhodopseudomonas capsulata upon variation of light intensity. Eur J Cell Biol. 1980 Dec;23(1):1–5. [PubMed] [Google Scholar]
- Hamana K., Kamekura M., Onishi H., Akazawa T., Matsuzaki S. Polyamines in photosynthetic eubacteria and extreme-halophilic archaebacteria. J Biochem. 1985 Jun;97(6):1653–1658. doi: 10.1093/oxfordjournals.jbchem.a135223. [DOI] [PubMed] [Google Scholar]
- Hiraishi A., Masamune K., Kitamura H. Characterization of the bacterial population structure in an anaerobic-aerobic activated sludge system on the basis of respiratory quinone profiles. Appl Environ Microbiol. 1989 Apr;55(4):897–901. doi: 10.1128/aem.55.4.897-901.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOVACS N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 1956 Sep 29;178(4535):703–703. doi: 10.1038/178703a0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Pagel J. E., Seyfried P. L. Numerical taxonomy of aquatic Acinetobacter isolates. J Gen Microbiol. 1976 Aug;96(2):220–232. doi: 10.1099/00221287-95-2-220. [DOI] [PubMed] [Google Scholar]