Abstract
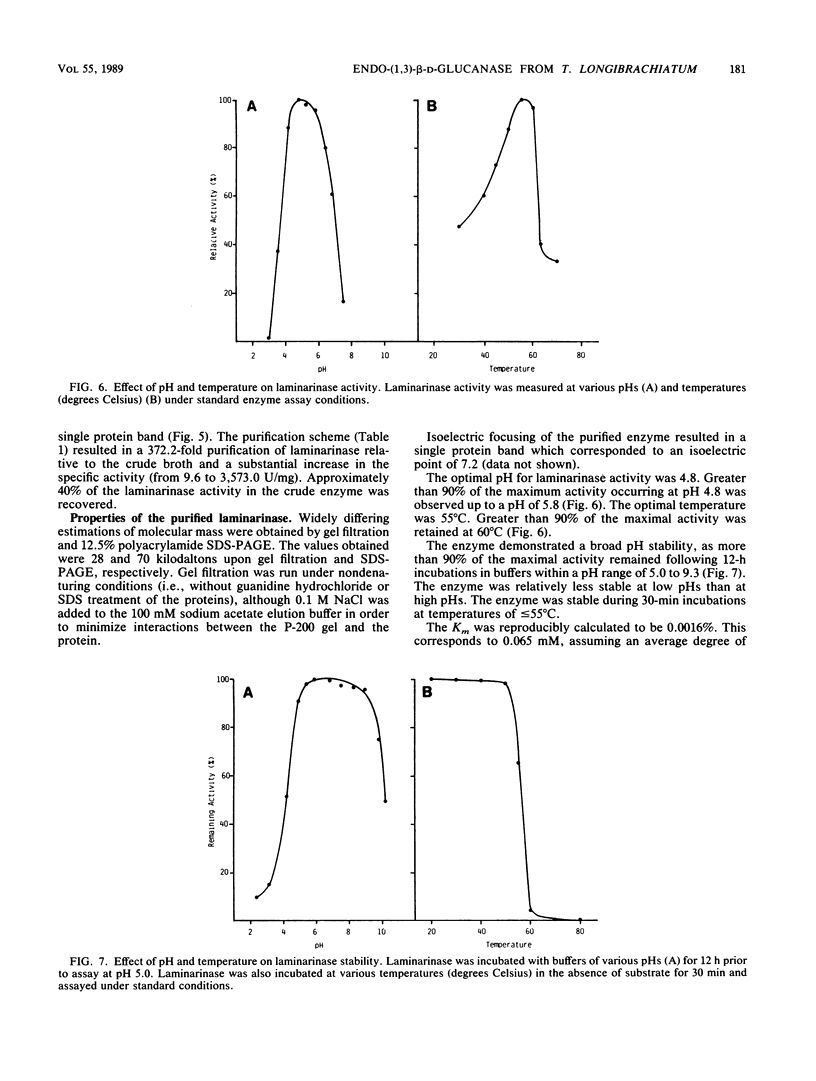
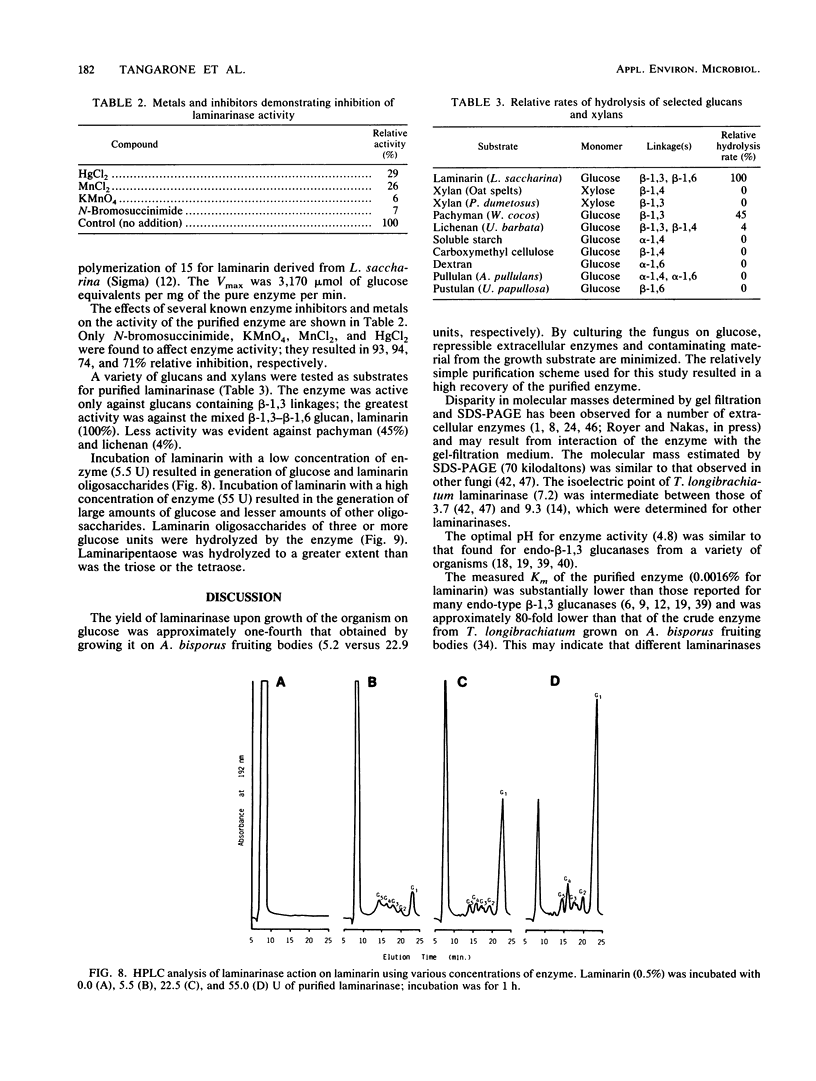
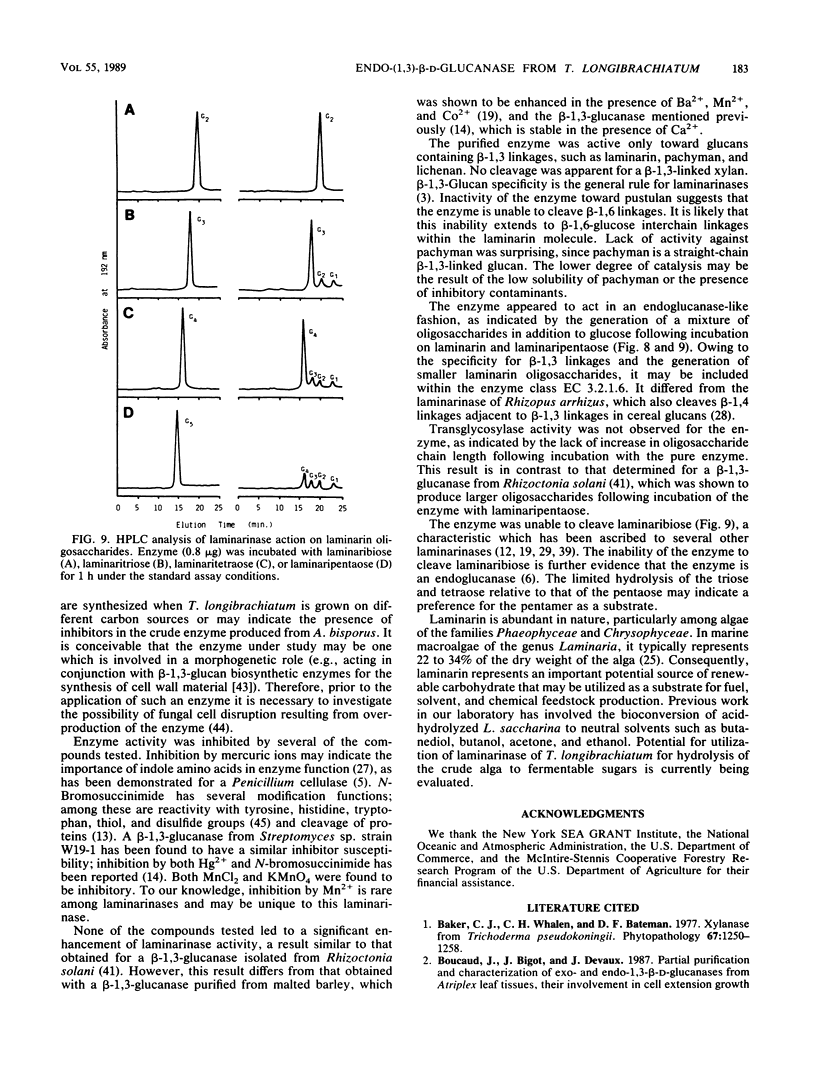
A laminarinase [endo-(1,3)-β-d-glucanase] has been purified from Trichoderma longibrachiatum cultivated with d-glucose as the growth substrate. The enzyme was found to hydrolyze laminarin to oligosaccharides varying in size from glucose to pentaose and to lesser amounts of larger oligosaccharides. The enzyme was unable to cleave laminaribiose but hydrolyzed triose to laminaribiose and glucose. The enzyme cleaved laminaritetraose, yielding laminaritriose, laminaribiose, and glucose, and similarly cleaved laminaripentaose, yielding laminaritetraose, laminaritriose, laminaribiose, and glucose. The enzyme cleaved only glucans containing β-1,3 linkages. The pH and temperature optima were 4.8 and 55°C, respectively. Stability in the absence of a substrate was observed at temperatures up to 50°C and at pH values between 4.9 and 9.3. The molecular mass was determined to be 70 kilodaltons by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis, and the pI was 7.2. Enzyme activity was significantly inhibited in the presence of HgCl2, MnCl2, KMnO4, and N-bromosuccinimide. The Km of the enzyme on laminarin was 0.0016%, and the Vmax on laminarin was 3,170 μmol of glucose equivalents per mg of the pure enzyme per min.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bull A. T., Chesters C. G. The biochemistry of laminarin and the nature of laminarinase. Adv Enzymol Relat Areas Mol Biol. 1966;28:325–364. doi: 10.1002/9780470122730.ch5. [DOI] [PubMed] [Google Scholar]
- Eriksson K. E., Pettersson G. Studies on cellulolytic enzymes. V. Some structural properties of the cellulase from Penicillium notatum. Arch Biochem Biophys. 1968 Mar 20;124(1):160–166. doi: 10.1016/0003-9861(68)90316-0. [DOI] [PubMed] [Google Scholar]
- Fleet G. H., Phaff H. J. Glucanases in Schizosaccharomyces. Isolation and properties of the cell wall-associated beta(1 leads to 3)-glucanases. J Biol Chem. 1974 Mar 25;249(6):1717–1728. [PubMed] [Google Scholar]
- Giulian G. G., Moss R. L., Greaser M. Analytical isoelectric focusing using a high-voltage vertical slab polyacrylamide gel system. Anal Biochem. 1984 Nov 1;142(2):421–436. doi: 10.1016/0003-2697(84)90486-x. [DOI] [PubMed] [Google Scholar]
- Jones D., Gordon A. H., Bacon J. S. Co-operative action by endo- and exo-beta-(1 leads to 3)-glucanases from parasitic fungi in the degradation of cell-wall glucans of Sclerotinia sclerotiorum (Lib.) de Bary. Biochem J. 1974 Apr;140(1):47–55. doi: 10.1042/bj1400047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronman M. J., Robbins F. M., Andreotti R. E. Reaction of N-bromosuccinimide with lysozyme. Biochim Biophys Acta. 1967 Dec 12;147(3):462–472. doi: 10.1016/0005-2795(67)90006-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manners D. J., Masson A. J., Patterson J. C. The structure of a beta-(1 leads to 3)-D-glucan from yeast cell walls. Biochem J. 1973 Sep;135(1):19–30. doi: 10.1042/bj1350019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners D. J., Wilson G. Purification and properties of an endo-(1 leads to 3)-beta-D-glucanase from malted barley. Carbohydr Res. 1974 Oct;37(1):9–22. doi: 10.1016/s0008-6215(00)87061-5. [DOI] [PubMed] [Google Scholar]
- Manners D. J., Wilson G. Studies on beta-glucanases. Some properties of a bacterial endo-beta-(1 leads to 3)-glucanase system. Biochem J. 1973 Sep;135(1):11–18. doi: 10.1042/bj1350011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen B. L., Brown L. R. The basis for colored silver-protein complex formation in stained polyacrylamide gels. Anal Biochem. 1984 Sep;141(2):311–315. doi: 10.1016/0003-2697(84)90047-2. [DOI] [PubMed] [Google Scholar]
- Paice M. G., Jurasek L., Carpenter M. R., Smillie L. B. Production, characterization, and partial amino acid sequence of xylanase A from Schizophyllum commune. Appl Environ Microbiol. 1978 Dec;36(6):802–808. doi: 10.1128/aem.36.6.802-808.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott W. A., Chapman J. M. A new improved method for the assay of endo-beta-1,3-glucanase. Anal Biochem. 1977 May 1;79(1-2):257–262. doi: 10.1016/0003-2697(77)90400-6. [DOI] [PubMed] [Google Scholar]
- RAMACHANDRAN L. K., WITKOP B. THE INTERACTION OF MERCURIC ACETATE WITH INDOLES, TRYPTOPHAN, AND PROTEINS. Biochemistry. 1964 Nov;3:1603–1611. doi: 10.1021/bi00899a001. [DOI] [PubMed] [Google Scholar]
- REESE E. T., MANDELS M. Beta-D-1, 3 Glucanases in fungi. Can J Microbiol. 1959 Apr;5(2):173–185. doi: 10.1139/m59-022. [DOI] [PubMed] [Google Scholar]
- Rombouts F. M., Phaff H. J. Lysis of yeast cell walls. Lytic beta-(1 leads to 3)-glucanases from Bacillus circulans WL-12. Eur J Biochem. 1976 Mar 16;63(1):121–130. doi: 10.1111/j.1432-1033.1976.tb10214.x. [DOI] [PubMed] [Google Scholar]
- SMOGYI M. Notes on sugar determination. J Biol Chem. 1952 Mar;195(1):19–23. [PubMed] [Google Scholar]
- Scott J. H., Schekman R. Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol. 1980 May;142(2):414–423. doi: 10.1128/jb.142.2.414-423.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallenberger R. S., Searles C., Lewis B. A. Laminaranase activity in the crystalline style of the surf clam (Spissula solidissima). Experientia. 1974 Jun 15;30(6):597–598. doi: 10.1007/BF01921493. [DOI] [PubMed] [Google Scholar]
- Skujins J. J., Potgieter H. J., Alexander M. Dissolution of fungal cell walls by a streptomycete chitinase and beta-(1-3) glucanase. Arch Biochem Biophys. 1965 Aug;111(2):358–364. doi: 10.1016/0003-9861(65)90197-9. [DOI] [PubMed] [Google Scholar]
- WITKOP B. Nonenzymatic methods for the preferential and selective cleavage and modification of proteins. Adv Protein Chem. 1961;16:221–321. doi: 10.1016/s0065-3233(08)60031-5. [DOI] [PubMed] [Google Scholar]
- Wong Y. S., Maclachlan G. A. 1,3-beta-D-glucanases from Pisum sativum seedlings. I. Isolation and purification. Biochim Biophys Acta. 1979 Dec 7;571(2):244–255. doi: 10.1016/0005-2744(79)90095-0. [DOI] [PubMed] [Google Scholar]
- del Rey F., Santos T., García-Acha I., Nombela C. Synthesis of 1,3-beta-glucanases in Saccharomyces cerevisiae during the mitotic cycle, mating, and sporulation. J Bacteriol. 1979 Sep;139(3):924–931. doi: 10.1128/jb.139.3.924-931.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]