Abstract
Evaluation of potential antineoplastic therapies would be enhanced by noninvasive detection of tumor cells in living animals. Because light is transmitted through mammalian tissues, it was possible to use bioluminescence to monitor (both externally and quantitatively) growth and regression of labeled human cervical carcinoma (HeLa) cells engrafted into immunodeficient mice. The efficacy of both chemotherapy and immunotherapeutic treatment with ex vivo expanded human T cell-derived effector cells was evaluated. In the absence of therapy, animals showed progressive increases in signal intensity over time. Animals treated with cisplatin had marked reductions in tumor signal; 5′-fluorouracil was less effective, and cyclophosphamide was ineffective. Immunotherapy dramatically reduced signals at high effector-to-target cell ratios, and significant decreases were observed with lower ratios. This model system allowed sensitive, quantitative, real-time spatiotemporal analyses of the dynamics of neoplastic cell growth and facilitated rapid optimization of effective treatment regimens.
Developments in cancer therapy have led to impressive improvements in disease-free survival; however, effective intervention strategies that target the small numbers of neoplastic cells remaining after therapy or early in the course of disease are major treatment challenges (1, 2). A significant percentage of patients who have had an apparent complete response to therapy enter a state of minimal residual disease that will eventually result in relapse. In addition, as the sensitivity of diagnostic tests improve, patients will be identified earlier in the course of their disease, either during a relapse or at initial diagnosis, preferably at a time when tumor load is minimal. The development and optimization of therapies designed to treat patients with reduced or limited tumor-cell burden may significantly enhance the overall results of therapeutic interventions.
Advancements in the treatment of minimal residual disease states have been limited by a lack of adequate animal models in which a small number of tumor cells can be detected and the patterns of growth and distribution can be studied over time. Mice with severe combined immunodeficiency (SCID) have been useful for studying human tumor xenografts (3–5). In some situations, the ability of the tumor cells to grow in SCID mice has been a powerful predictor of the clinical course of the patient (6). However, detection of neoplastic cells in current animal models of human disease requires a large number of target cells, and such models typically use the therapeutic endpoints of gross tumor growth or death of the animal. Molecular techniques, such as DNA amplification by using PCR, have the sensitivity to detect DNA targets from small numbers of transformed cells but are hampered by sampling limitations and often require killing of the animal subjects (7, 8). Although a variety of other surrogate markers of tumor growth have been used in animal models (9), noninvasive, direct, and sensitive quantification of tumor burden in vivo has not been shown previously. Monitoring tumor growth, metastases, and response to therapeutic interventions in animal models of minimal disease states are critical for the development of effective strategies that target small numbers of neoplastic cells.
To enable studies of minimal disease, we have developed a sensitive imaging approach that can be used to detect quantitatively small numbers of cells and, through the use of whole-body imaging, follow tumor burden over several logs of cell growth. This technique was accomplished by introducing a constitutively expressed bioluminescent reporter gene encoding the photoprotein firefly luciferase into the chromosomes of a tumor-cell line, transferring small numbers of these labeled cells into immunodeficient mice, and monitoring the distribution and growth of these cells by using the light produced by the transformed cells and transmitted through the anesthetized animals’ tissues to monitor the distribution and growth of tumor cells (10). The use of this method for the study of infection and gene expression in living animals has been demonstrated effectively in earlier studies (11–13).
Using this methodology, we have evaluated both conventional chemotherapies and an immune cell therapy that uses cytokine-induced killer (CIK) T cells. CIK cells are generated by culturing peripheral blood mononuclear cells in the presence of IFN-γ, followed 1 day later by IL-2 and an anti-CD3 monoclonal antibody (14–16). This incubation results in the dramatic expansion of effector cells that have cytolytic activity against a broad array of tumor-cell targets, including multidrug resistant cell lines and autologous fresh tumor isolates (17–19). The cells with the most robust antitumor activity express CD3, a T cell marker, and the natural killer cell marker CD56. Effector cells with this phenotype in the peripheral blood have also been shown to have cytolytic activity (20–22). Cell killing by these effector cells seems to proceed through a non-MHC-restricted mechanism (17, 23). Currently, CIK cells are being evaluated in a phase I/II clinical trial as adoptive immunotherapy for patients with recurrent lymphoma and Hodgkin’s disease.
In this study, conventional chemotherapies and adoptive immunotherapy with human CIK cells were evaluated and compared by monitoring spatiotemporal distribution of bioluminescent human-derived tumor cells in living immunodeficient mice. In the absence of therapy, there were progressive increases in signal intensities as an indication of increasing tumor burden. In contrast, significant reductions in signal intensities were observed in the treatment groups. This model system allowed sensitive, real-time analyses of the dynamics and quantification of neoplasia in time and space and facilitated rapid optimization of effective treatment regimens.
Methods
Transfection of Tumor-Cell Lines.
The human cervical carcinoma line HeLa was transfected with a modified firefly luciferase gene (pGL3 series of vectors, Promega), and stable lines were generated under Zeocin (Invitrogen) selection. The pGL3 control plasmid and the pZeo-SV2 vector (Invitrogen), which confers resistance to the bleomycin analog Zeocin, were cotransfected into HeLa cells by using the liposome formulation Lipofectamine (GIBCO/BRL) according to the manufacturer’s recommendations. Optimal conditions for DNA delivery were identified by adding luciferin (0.5 μg/ml final; Molecular Probes) to the cell culture medium. Selection of stable transfectants was performed in Zeocin (500 μg/ml), and light emission was used to confirm expression of the reporter gene. Cultures were screened by using an intensified charge-coupled device camera (C2400-32, Hamamatsu Photonics, Hamamatsu City, Japan). Colonies of cells expressing light were grown in DMEM supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml), glutamine (2 mmol/liter), 2-mercaptoethanol (50 μmol/liter), FCS [10% (vol/vol); HyClone], and Zeocin (500 μg/ml).
Imaging of Tumor Xenografts.
Before imaging, mice were anesthetized with pentobarbital (70 mg/kg body weight). An aqueous solution of the substrate luciferin (Molecular Probes; 50 mM; 126 mg/kg) was injected into the peritoneal cavity 5 min before imaging. The animals were then placed in a light-tight chamber, and a gray-scale body-surface reference image was collected with the chamber door slightly open. For this purpose, a low-light imaging system, comprised of an intensified charge-coupled device camera fitted with a 50-mm f1.2 Nikkor lens (Nikon) and a computer with image-analysis capabilities, was used (10–13). Subsequently, the door to the chamber was closed to exclude the room light that obscured the relatively dimmer luciferase bioluminescence. Photons emitted from luciferase within the animal and then transmitted through the tissue were collected and integrated for a period of 5 min. A pseudocolor image representing light intensity (blue least intense and red most intense) was generated on an Argus 20 image processor (Hamamatsu); images were transferred by using a plug-in module (Hamamatsu) to a computer (Macintosh 8100/100) running an image-processing application (photoshop, Adobe Systems, Mountain View, CA). Gray-scale reference images and pseudocolor images were superimposed by using the image-processing software, and annotations were added by using another graphics software package (canvas, version 5.0, Deneba, Miami, FL).
Generation of CIK Cells.
CIK cells were prepared from human peripheral blood mononuclear cells as described (15). Cells were cultured in complete medium consisting of RPMI medium 1640 (Applied Scientific, San Francisco), 10% (vol/vol) FCS (HyClone), 25 mmol/ml penicillin, 100 μg/ml streptomycin, 2 mmol/liter l-glutamine, and 50 μmol/liter 2-mercaptoethanol. Human recombinant IFN-γ (Genentech) was added at a concentration of 1,000 units/ml. After 24 h of incubation, 50 ng/ml of CD3 (OKT3, Ortho Biotec, Raritan, NJ) and 300 units/ml human recombinant IL-2 (Chiron) were added. Cells were subcultured every 5 days in fresh complete medium containing 300 units/ml of IL-2. In vitro cytotoxicity against OCI-Ly8 (B cell non-Hodgkin’s lymphoma), HeLa, and HeLa-luc cells was assessed by cytotoxicity assays as described (14).
PCR.
Cells from the peritoneal cavities of mice that had been killed were removed by lavage by using 10 ml of ice-cold RPMI medium 1640. Genomic DNA was prepared by using a phenol-chloroform extraction. DNA from 1 × 107 cultured mouse cells (NIH 3T3 alone or a serial dilution of HeLa with a stable integration of the luciferase gene) was extracted. As a marker for the xenograft, the luciferase gene was amplified by using a nested set of primers from the prepared genomic DNA. The 5′ and 3′ primers were RL1 (5′-agtAAGCTTGGCATTCCGGTACTGTTGGTAAAGCCACCATGGAAG-3′ at positions 245–286 of pGL3-control plasmid; Promega) and LucR600 (5′-GACGAACGTGTACATCGAGTGAAATCCCTGGTAAT-3′ at positions 749–783). PCRs were carried out in a buffer system (10 mM Tris⋅HCl, pH 8.3/50 mM KCl/1% DMSO) for 3 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min and 37 cycles of 94°C for 10 s, 60°C for 20 s, and 72°C for 2 min in the presence of 1.75 mM MgSO4, 200 μM of each dNTP, 10 pmol of each primer, 1 μg of genomic DNA, and 2.5 units of Taq polymerase (24). The second-round primers were Luctop (5′-ttaggatccGAAGACGCCAAAAACATAAAGAAA-3′ at positions 283–306) of pGL3 and RL6seq (5′-CCCCTTTTTGGAAACGAACACCACGGTAG-3′ at positions 647–675). The template for the second-round reactions consisted of 1 μl of a 1–10 dilution of the first-round product. Second-round reactions were performed with 3 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 2 min and 37 cycles of 94°C for 10 s, 60°C for 20 s, and 72°C for 2 min. The control PCR for mouse genomic DNA was performed for 3 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 2 min and 37 cycles of 94°C for 10 s, 57°C for 20 s, and 72°C for 2 min by using primers Hsp70.2top (5′-ttggagctcAGTCAGTGGCGGCACCCGACAC-3′ at positions −619 to −598 from ATG, the translational start of the hsp70.2 gene) and Hsp70.2bot (5′-cagccatggTGACTGAAAAAACAGGCAAAGCAACGTT-3′ at positions −30 to −3 from ATG), which produces a 633-bp DNA fragment from the promoter region of the gene encoding a 70-Kd heat-shock protein (Hsp70).
Statistical Methods.
For both sets of chemotherapy and immunotherapy comparisons, the logarithm of each tumor signal was used, comparing the geometric means at each time point. For each set of experiments, ANOVA was first performed, each yielding statistically significant differences among experimental groups at the 0.05 level. The t test was then used to test the difference between each experimental group and the control group, and their nominal P values are reported. To adjust for multiple comparisons that used the same control in each of the experiments, the Dunnett’s test was also used (25).
Results
Sensitivity of Detection.
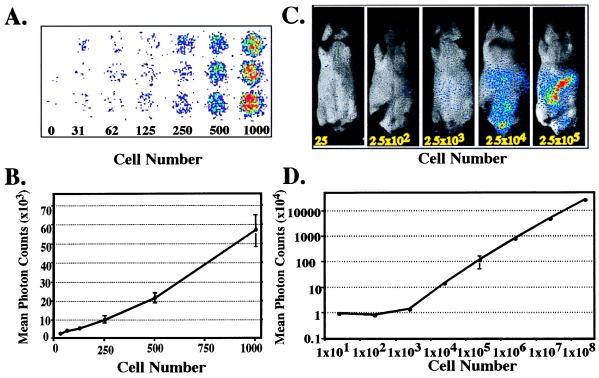
The human-derived HeLa cell line was labeled through expression of a stable integrant of a modified luciferase (luc) gene derived from the North American firefly (Photinus pyralis). These labeled cells are referred to as HeLa-luc cells. The sensitivity of detecting HeLa-luc cells in vitro culture is shown in Fig. 1A. As few as 30 cells can be detected above background, and their signal can be quantitated (Fig. 1B). To determine the sensitivity of detection in vivo, logarithmic dilution’s of HeLa-luc cells were injected into the peritoneal cavity of SCID mice that had received prior irradiation of 200 cGy to limit endogenous natural killer cell activity (26). The light generated internally by the photoprotein expressed in the tumor cells and then transmitted through the animals’ tissues was monitored externally by using an intensified charge-coupled device camera (13). The substrate luciferin was provided exogenously via i.p. injection. In this xenograft model of human neoplasia, as few as 2,500 tumor cells distributed throughout the peritoneal cavity could be detected externally (Fig. 1C). Quantitation was achieved by integrating the signal intensity over time and was reported as total photon counts. The photon counts externally detected from the animals were proportional to the input number of cells injected over 4 logs of tumor cells (Fig. 1D). Because quantitation of total photons, as an indicator of tumor burden, was performed in living animals, progressive tumor growth or regression could be monitored by repeated analyses of animals in a single group at serial time points.
Figure 1.
Sensitivity of detection of HeLa-luc cells in culture and in animals. (A and B) Dilution series of HeLa-luc cells were imaged in 96-well cell culture plates over a range of 0–1,000 cells. Photon counts from each of the triplicate wells were determined with a 5-min integration time, and the means of total photon counts were plotted with respect to cell number. (C) CB-17 SCID mice were irradiated with 200 cGy to eradicate residual natural killer activity. HeLa-luc cells, in a series ranging from 25 cells to 2.5 × 108 cells, were injected into the mice (i.p.). Groups of 3–6 mice at each cell concentration (except for 2.5 × 108 cells, which represents a single animal) were used to establish the limits of detection in vivo. Only the animals injected with the fewest cells are displayed here to show the sensitivity of the detection method. (D) Signals from the entire abdominal region of each mouse were quantified, and the mean photon counts were plotted. There was limited variation for each of the data points, which was <0.1% for all data points, with the exception of 2.5 × 105 cells, which was <5%.
Response to Chemotherapy in Vivo.
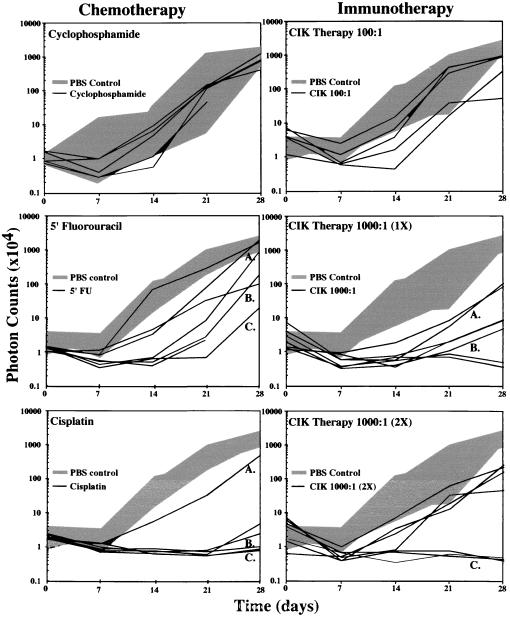
Temporal analyses of tumor-cell clearance after antineoplastic therapies in vivo were performed in SCID mice engrafted with HeLa-luc cells. The kinetics of tumor-cell growth were evaluated in groups of mice that were treated with i.p. injections of cyclophosphamide (1 mg/kg per day on days 2 and 3), 5′-fluorouracil (5′-FU; 25 mg/kg per day on days 2, 4, 6, and 8), cisplatin (3 mg/kg per day on days 2, 5, and 8), or PBS (1 ml per animal on day 1 as untreated control). Mice were imaged weekly for 4 weeks, and total numbers of photons transmitted from the HeLa-luc cells in vivo were determined (Fig. 2). Detecting early indications of therapeutic efficacy well before the appearance of the traditional endpoints of weight loss, palpable tumor nodules, or death was possible in these analyses. In addition, it was possible to evaluate individual trajectories of tumor-cell growth over time in these mice, and thus, patterns of cell growth were revealed in living animals without the limitations associated with tissue sampling or using death as an end point.
Figure 2.
Tumor-cell growth in animals with and without therapy. SCID mice, 6–8 weeks old, were irradiated with 200 cGy to eradicate functional natural killer cell activity (day 0). The following day, the mice received i.p. injections of 1 × 104 HeLa-luc cells, and baseline images and intensities were obtained. The mice were then treated with cyclophosphamide, 5′-FU, or PBS on day 1 as control. Tumor progression was monitored by measuring light emission from individual mice every 7 days for a period of 28 days. Growth of tumors in individual treated mice is shown with black lines, and the range of tumor growth in the absence of therapy is shown by the gray area. Growth curves designated by letters (A–C) indicate those tumor-growth profiles that show a range of therapeutic responses that were selected for presentation in Figs. 3 and 4. In the immunotherapy treatment groups, SCID mice received either CIK cells or PBS as control.
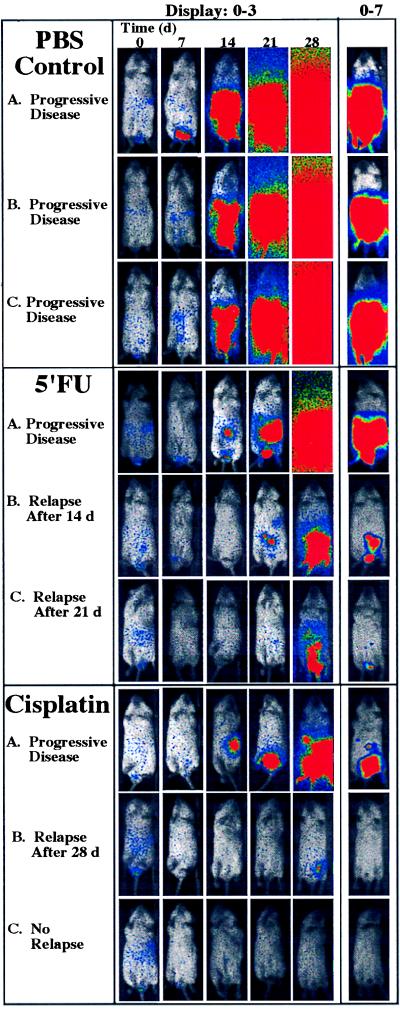
A range of bioluminescent patterns was apparent in each of the treatment groups. These ranged from apparently unimpeded exponential growth to inhibition of growth with subsequent relapse or reduction of signal below detectable levels for the entire study period (Fig. 3). Among mice treated with cyclophosphamide, there were no apparent differences in tumor growth compared with the control group. In contrast, mice treated with 5′-FU showed attenuation of signal from tumor cells initially; however, relapse was observed in each animal at various times, indicating a delay compared with untreated controls. Significant signal reductions were observed in the cisplatin-treated group (Fig. 2). The total photon counts obtained for several of the animals in this group were at or below background levels. Once reduced, these signals remained undetectable over the entire 28-day observation period. In mice treated with cisplatin, the tumor burden, as indicated by the bioluminescent signal, was significantly different from the control group at each of the four observation points in the experiment: at day 7 after tumor engraftment, the P value was 0.03; at day 14, P = 0.0001; at day 21, P = 0.0001; and at day 28, P = 0.0009. To account for multiple comparisons with a single control group, the Dunnett’s test was also used, resulting in statistical significance at the <0.05 level at each time point. Images of animals in this treatment group did reveal spatial distribution and temporal patterns of tumor growth representing a full range of response to therapy including clearance, relapse, and apparent unimpeded growth (Fig. 3). In all animals where a signal above background became apparent, tumor expansion ensued. During such relapses, initially, a focal pattern was observed that appeared to progress to a more generalized pattern of light emission. However, signals from anatomical sites other than the abdominal region never became apparent in any animal at any time. Thus, despite uniformity of inoculation and therapy, evidence of unique, individualized patterns of tumor regression and growth in the presence of therapy were apparent.
Figure 3.
Tumor dynamics after chemotherapy in vivo. Images representing the entire time course for selected animals in each treatment group are shown; letters correspond to plotted data designated A–C in Fig. 2. Three representative animals were selected from the PBS control group. Profiles of animals with tumor-growth trajectories representing the full range of therapeutic response are shown for the 5-FU and cisplatin groups. Images representing the full time course for each animal are displayed identically at a bit range of 0–3 (first five columns) to show the exponential increases indicated in Fig. 2. The last image, obtained at day 28, is displayed again at a bit range of 0–7 to reveal the anatomic localization of the signals (last column).
Response to Immunotherapy.
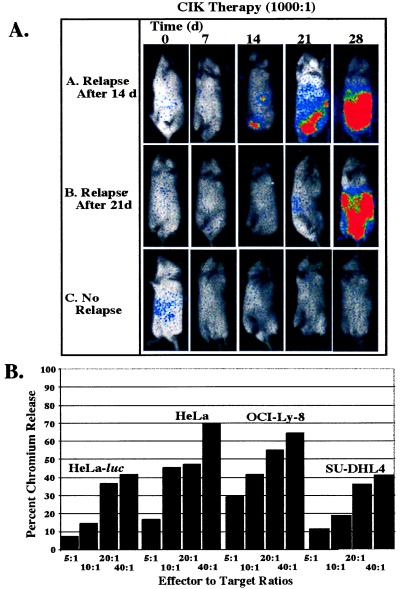
A primary objective of this study was to evaluate the effects of CIK cells on the in vivo growth of tumor cells in minimal residual disease states. To establish a benchmark of CIK cell cytotoxicity against selected targets, in vitro chromium release assays were performed by using either the HeLa human cervical cell line or the HeLa-luc derivative as targets and comparing these results to those obtained with the human B lymphoma cell lines OCI-Ly8 and SU-DHL4. Standardized killing between 40 and 80% was observed against all four target lines at an effector-to-target cell (E:T) ratio of 40:1, with HeLa-luc cells being marginally less sensitive to killing than the parental HeLa line (Fig. 4B). Previously, CIK cells have been shown to be effective in eradicating the two human B cell lymphoma cell lines as well as autologous chronic myelogenous leukemia lines in SCID mouse models (14, 18). These in vitro cytotoxicity assays suggested that expression of a foreign protein in the form of luciferase did not affect killing profiles, and thus, labeled cell lines may also be similarly susceptible to CIK cell killing in vivo.
Figure 4.
CIK immunotherapy in vivo and response of cell lines to CIK cells in vitro. (A) Time courses for animals were selected to show the full spectrum of response to immunotherapy. Animals A and B show relapse of tumor growth after days 14 and 21, respectively. Animal C shows complete eradication of tumor signal throughout the 28-day experimental period. (B) In vitro cytotoxicity of CIK cells against HeLa was not altered by luciferase expression. Other cell lines are shown as representative targets for CIK cells.
The in vivo activity of CIK cells against the HeLa-luc cell line was then studied, and the results were compared with the response of HeLa-luc cells to chemotherapeutic agents. Three immunotherapy treatment groups were evaluated following i.p. injection of 1 × 104 HeLa-luc cells. A representative experiment is shown in Fig. 2. Groups of seven mice received two treatments of 1 × 106 CIK cells (E:T ratio of 100:1) on days 1 and 7, a single dose of 1 × 107 CIK cells (E:T ratio of 1,000:1) on day 1, or two doses of 1 × 107 CIK cells (E:T ratio of 1,000:1) on days 1 and 7. A fourth group received PBS only. All animals that received CIK cell therapy had statistically significant reductions in tumor signal compared with PBS-control animals (Figs. 2 and 4). The group receiving 1 × 106 CIK cells showed a trend toward attenuation of tumor signal early in the treatment period (P = 0.2 at day 7), and a significant difference was observed by day 21 (P = 0.01). By 28 days after treatment, significant differences from the control group were not apparent, and all of the mice in this group had detectable tumor signals. In contrast, the groups receiving one or two treatments of 1 × 107 CIK cells showed considerable reduction of tumor signal. At the end of the 28-day treatment period, two of six mice treated with a single dose of CIK cells were free of disease, and an additional two animals had markedly reduced tumor signals as assessed by light emission. In animals that received two doses of CIK cells, three of seven animals were completely free of disease, whereas other animals had a reduction in tumor signal, although tumor growth was detectable (Fig. 4A). Statistically significant differences in the mean values of tumor signals could be detected within 7 days (P = 0.01 for one CIK cell treatment of 107 cells, and P = 0.007 for two treatments of 107 cells). These differences persisted throughout the 28-day observation period. (On day 14, P = 0.0001; on day 21, P = 0.0001; and on day 28, P = 0.0004 for one treatment. On day 14, P = 0.0001; on day 21, P = 0.0012; and on day 28, P = 0.007 for two treatments.) In addition to the t tests, Dunnett’s test was also used to correct for multiple comparisons with a single control group. Statistical significance at the <0.05 level resulted at the same time points between the same groups as above. Despite diminished signal intensities in all CIK-treated animals, increased efficacy was observed at higher E:T ratios and with multiple treatments as indicated by the different patterns of tumor-cell growth and clearance.
PCR Analysis of Long-Term Survivors.
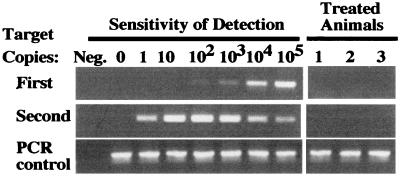
Long-term follow-up of the animals revealed survival differences between the control group and the groups treated either with CIK cells or cisplatin (P < 0.01). These survival data correlated well with the reductions in light intensity seen within these treatment groups during the first 4 weeks of the experiment. Long-term survivors had a complete absence of bioluminescent signals at >75 days after treatment. DNA from cells obtained by peritoneal lavage from three of the cisplatin-treated animals was evaluated by PCR to determine whether the tumor cells were detectable by another sensitive assay (Fig. 5). Despite sensitivity to approximately a single copy of the luciferase gene, no signal was detected in cellular DNA from long-term survivors, substantiating the results of the bioluminescence assay in which signals also were not apparent.
Figure 5.
PCR analyses of long-term survivors in the cisplatin-treated group. Mice were evaluated by PCR to determine whether DNA from the xenograft could be detected in the absence of a detectable bioluminescent signal. PCR with two sets of nested primers for the luciferase gene was performed on DNA from cells obtained from the peritoneal cavity.
Discussion
Improvements and modifications to animal model systems are needed to understand further the various mechanisms of immune containment of neoplasia and to develop therapies to prevent and treat small numbers of neoplastic cells present in minimal disease states (6). We addressed this previously unmet need by developing a functional imaging method that permits the study of small numbers of tumor cells in vivo in a noninvasive manner. As few as 2,500 tumor cells distributed throughout the peritoneal cavity could be reliably detected and quantitated in living animals. Light emission was proportional to tumor-cell inocula over a broad dynamic range of cell number over at least four orders of magnitude. Thus, the kinetics of tumor growth could be evaluated over time because of this broad dynamic range.
By using this method, real-time functional data indicating spatial distributions of tumor cells could be acquired at multiple times during the disease course. Therefore, single animals could be followed over time, removing intersample variability, with the net result of improved statistical analyses. An additional advantage of using cells labeled with a light-emitting optical signature such as luciferase for these analyses is that the molecular tag is copropagated with the target cells. Therefore, external signals are proportional to tumor-cell burden (over several logs) and do not decrease as the cell population increases as would occur with a contrast agent, radionuclide label, or vital dye that would decay or be diluted with increases in the cell numbers through cell division (26).
By using this approach, the effects of both chemotherapy and immunotherapies in the treatment of a human cervical carcinoma cell line were rapidly evaluated in SCID mice, revealing the in vivo dynamics of tumor regression and the efficacy of CIK cell, 5′-FU, and cisplatin therapies. Both CIK cell treatment and cisplatin therapy resulted in large reductions in tumor signal with good statistical significance beginning at 7–14 days and continuing throughout the study period. A dose-dependent response was apparent among the CIK cell-treated animals. For example, in the representative experiment shown in Fig. 2, the lower effector cell dosage (1 × 106 CIK cells) resulted in transient responses in three of five animals treated, in contrast to the animals treated with higher cell doses (1 × 107 cells for one or two treatments) that showed long-term reductions in signal intensities. As early as days 7 and 14, these responses were significantly different than those of untreated control animals. Approximately half of the animals receiving 1 × 107 effector cells had a complete resolution of tumor signals. Because the extent and tempo of disease can be monitored in intact living animals, this approach provides a “window” into early tumor-growth kinetics and response to therapy with the ability to follow disease progression for weeks. This model system allows for the rapid optimization of therapies directed toward the control of disease at times of minimal tumor burden. Furthermore, patterns of relapse and metastases can be followed, which may help to direct the choices of treatment modalities, combination therapies, and routes of administration.
Animal models traditionally have been cumbersome because of the difficulty in quantitating tumor burden and the requirement for either bulk tumor growth or animal survival as end points to evaluate the effect of a potential therapy (5). Our imaging strategy provides information concerning the spatial and temporal patterns of tumor-cell growth that can be accessed quickly. Thus, rapid assessment of a potential intervention in a fully quantitative manner is possible. Because far fewer animal subjects are required to obtain statistically meaningful results and because the data obtained reveal functional information, animal studies can be refined. Moreover, because the studies can be performed in minimal disease states, the stress on the animals that are studied can be reduced dramatically.
With this approach, it should be possible to track, not only the fate of tumor cells, but also the tissue distribution of effector cells and thus begin to analyze the mechanisms of immune surveillance of neoplasia in vivo. Theoretically, other cell populations or biological functions that can be suitably labeled are amenable for study in living hosts with human diseases. Further modifications of this model system with closely related photoproteins that have similar biochemical characteristics but different emission wavelengths may permit the development of dual-color assays for monitoring tumor progression and interaction of the host response in the same animals (27).
Acknowledgments
We thank Dr. Ruby M. Wong and Mr. Roger Baldwin for the statistical analyses. This work was supported in part by Public Health Service Grant CA-49605, a Translational Award from The Leukemia Society of America, funds from the Medical Free Electron Laser Program (ONR contract N-00014-94-1-1024), and unrestricted gifts from The Mary L. Johnson Fund, The Hess Research Fund, and The Baxter Foundation. We thank the Hamamatsu Corporation for technical support.
Abbreviations
- SCID
severe combined immunodeficiency
- CIK
cytokine-induced killer
- 5′-FU
5′-fluorouracil
- E:T ratio
effector-to-target cell ratio
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cavé H, van der Werff ten Bosch J, Suciu S, Guidal C, Waterkeyn C, Otten J, Bakkus M, Thielemans K, Grandchamp B, Vilmer E. N Engl J Med. 1998;339:591–598. doi: 10.1056/NEJM199808273390904. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch-Ginsberg C. Annu Rev Med. 1998;49:111–122. doi: 10.1146/annurev.med.49.1.111. [DOI] [PubMed] [Google Scholar]
- 3.Kamel-Reid S, Latarte M, Sirard C, Doedens M, Grunberger T, Fulop G, Freedman M H, Phillips R A, Dick J E. Science. 1989;246:1597–1600. doi: 10.1126/science.2595371. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi H, Nakada T, Puisieux I. Science. 1993;259:1460–1464. doi: 10.1126/science.8451642. [DOI] [PubMed] [Google Scholar]
- 5.Uckun F M. Blood. 1996;88:1135–1146. [PubMed] [Google Scholar]
- 6.Uckun F M, Sather H, Reaman G, Shuster J, Land V, Trigg M, Gunther R, Chelstrom L, Bleyer A, Gaynon P. Blood. 1995;85:873–878. [PubMed] [Google Scholar]
- 7.Zhang L, Hellström K E, Chen L. Clin Exp Metastasis. 1994;12:87–92. doi: 10.1007/BF01753974. [DOI] [PubMed] [Google Scholar]
- 8.Negrin R S, Blume K G. Blood. 1991;78:255–258. [PubMed] [Google Scholar]
- 9.Arguello F, Sterry J A, Zhao Y Z, Alexander M R, Shoemaker R H, Cohen H J. Blood. 1996;87:4325–4332. [PubMed] [Google Scholar]
- 10.Edinger M, Sweeney T S, Tucker A A, Olomu A B, Negrin R S, Contag C H. Neoplasia. 1999;1:303–310. doi: 10.1038/sj.neo.7900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contag C H, Contag P R, Mullins J I, Spilman S D, Stevenson D K, Benaron D A. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 12.Contag C, Spilman S, Contag P, Oshiro M, Eames B, Dennery P, Stevenson D, Benaron D. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 13.Contag P R, Olomu I N, Stevenson D K, Contag C H. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt-Wolf I G H, Negrin R S, Kiem H P, Blume K G, Weissman I L. J Exp Med. 1991;174:139–149. doi: 10.1084/jem.174.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu P H, Negrin R S. J Immunol. 1994;153:1687–1696. [PubMed] [Google Scholar]
- 16.Ochoa A C, Gromo G, Alter B J, Sondel P M, Bach F H. J Immunol. 1987;138:2728–2733. [PubMed] [Google Scholar]
- 17.Schmidt-Wolf I G, Lefterova P, Mehta B A, Fernandez L P, Huhn D, Blume K G, Weissman I L, Negrin R S. Exp Hematol. 1993;21:1673–1679. [PubMed] [Google Scholar]
- 18.Schmidt-Wolf I G, Lefterova P, Johnston V, Scheffold C, Csipai M, Mehta B A, Tsuruo T, Huhn D, Negrin R S. Cell Immunol. 1996;169:85–90. doi: 10.1006/cimm.1996.0094. [DOI] [PubMed] [Google Scholar]
- 19.Hoyle C, Bangs C D, Chang P, Kamel O, Mehta B, Negrin R S. Blood. 1998;92:3318–3327. [PubMed] [Google Scholar]
- 20.Schmidt R E, Murray C, Daley J F, Schlossman S F, Ritz J. J Exp Med. 1986;164:351–356. doi: 10.1084/jem.164.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanier L L, Le A M, Civin C I, Loken M R, Phillips J H. J Immunol. 1986;136:4480–4486. [PubMed] [Google Scholar]
- 22.Ortaldo J R, Winkler-Pickett R T, Yagita H, Young H A. Cell Immunol. 1991;136:486–495. doi: 10.1016/0008-8749(91)90369-m. [DOI] [PubMed] [Google Scholar]
- 23.Mehta B A, Schmidt-Wolf I G, Weissman I L, Negrin R S. Blood. 1995;86:3493–3499. [PubMed] [Google Scholar]
- 24.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rübsamen-Waigmann H, Mullins J I. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 25.Glantz S A, Slinker B K. Primer of Applied Regression and Analysis of Variance. New York: McGraw-Hill; 1990. [Google Scholar]
- 26.Shpitz B, Chambers C A, Singhal A B, Hozumi N, Fernandes B J, Roifman C M, Weiner L M, Roder J C, Gallinger S. J Immunol Methods. 1994;169:1–15. doi: 10.1016/0022-1759(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 27.Hendrikx P J, Martens C M, Hagenbeek A, Keij J F, Visser J W. Exp Hematol. 1996;24:129–140. [PubMed] [Google Scholar]
- 28.Kajiyama N, Nakano E. Protein Eng. 1991;4:691–693. doi: 10.1093/protein/4.6.691. [DOI] [PubMed] [Google Scholar]