Abstract
Herpes simplex virus 1 encodes several functions to preclude the shutoff of host response to infection, including degradation of mRNA immediately after infection. To determine whether any cellular mRNAs accumulate in infected cells against a background of severe loss of host RNA, we hybridized cDNAs derived from three different cell lines infected with wild type and a mutant virus to a DNA array containing probes for 588 human genes representing different functional groups. The results were that (i) infected cells accumulated at levels above those of mock-infected cells, a small number of transcripts representing transcriptional factors that could regulate gene expression both positively and negatively, and one stress response protein (GADD45), (ii) the amount and nature of the accumulated transcripts showed limited variability depending on the cell and virus, and (iii) at least some of the proteins encoded by the accumulated transcripts could benefit either the virus or the host.
Keywords: DNA arrays, GADD45
In productive infection of susceptible cells, the herpes simplex virus 1 (HSV-1) multiplies and ultimately destroys the cell (reviewed in ref. 1). Studies of the functions of viral gene products suggest that a considerable fraction of viral genetic information is designed to silence the infected cell and prevent it from responding to the presence of the virus and its gene products. In fact, the knowledge as to how the cell can respond has to a large extent become apparent from the functions HSV-1 evolved to silence it. In brief, the virus appears to have evolved three different sets of functions to suppress the cell.
The function of the first set is to preclude de novo cellular gene expression by blocking the maturation or by degradation of cellular RNA. This manifestation of infected cells is caused by at least two viral gene products. Thus the viral UL41 gene product packaged in the virion and known also as the virion host shutoff (vhs) protein causes massive indiscriminant degradation of mRNA (2–6). Although viral mRNA also is degraded, the sheer bulk of viral RNA transcripts insures that viral proteins are made. In addition, the infected cell protein no. 27 (ICP27), the product of the α27 gene, inhibits the function of cellular spliceosomes and blocks the maturation of cellular mRNA (7–9). This function is effective because only four viral proteins are known to be the products of spliced mRNA, and three of these are made immediately after infection presumably before ICP27 is fully effective (1).
The second set of viral functions precludes activation of preexisting cellular defense mechanism. Thus presentation of antigenic peptides is blocked by the binding of ICP47 to TAP1/TAP2 proteins whereas ICP34.5, the product of the γ134.5 gene, nullifies the effect of activation of protein kinase R (PKR) (10–14). Activation of PKR by double-stranded RNA results in the phosphorylation of the α subunit of the translation initiation factor 2 and complete shutoff of protein synthesis. ICP34.5 precludes the shutoff of protein synthesis by binding to and redirecting the cellular protein phosphatase 1 to dephosphorylate the translation initiation factor. In the example cited above, the virus sequesters for its own use a preexisting cellular protein to block host response to infection (13, 14).
The third set of viral functions preclude apoptosis induced in infected cells at one or more stages during the viral reproductive cycle (15, 16).
The overall pattern of decreased synthesis of cellular proteins does not imply that the host does not attempt to respond by de novo gene expression or that the virus does not induce specific accumulation of transcripts encoding desirable cellular proteins. The purpose of the studies described in this report was 2-fold. First, it was of interest to test the hypothesis that notwithstanding the massive shutoff of host macromolecular synthesis, the cell is able to respond or is induced to make a restricted set of cellular gene products. Second, because the γ134.5 gene blocks the ability of the cell to respond to infection and enable viral replication and spread in experimental animal systems, it was of interest to determine whether the host response to infection in the absence of the γ134.5 gene was significantly higher than in its presence (17).
In these studies we used DNA arrays containing a limited number of probes to test the accumulation of cellular transcripts in three different human cell lines. The human embryonic lung fibroblasts are the closest we could come to susceptible “normal” human cells in vitro. The two isogenic cancer cell lines differ with respect to the function of p53 and could provide information on the role of this gene in the course of infection. We report that in a background of significant decrease in overall accumulation of host mRNA, transcripts arising from a small fraction of the probed cellular genes accumulated in large amounts and that in a few instances the accumulation was both cell type and virus dependent.
Materials and Methods
Cells and Viruses.
Human lung fibroblasts (HLF) were maintained as described (18). U87-lux.8 and U87–175.4 are isogenic cell lines derived from the human U87MG glioma cell line (19). U87–175.4 cells express a dominant negative p53 mutant (codon 175arg→his) that interferes with wild-type p53 function. U87-lux.8 cells are stably transformed with the vector alone and an antibiotic resistance gene and are wild type for p53 function. Both isogenic cell lines were maintained in DMEM (GIBCO/BRL) supplemented with 10% heat-inactivated FCS, nonessential amino acids, and 400 μg/ml of G418. HSV-1(F) is the prototype HSV-1 strain used in our laboratories. The R3616 recombinant virus was derived from HSV-1(F) by deletion mutagenesis of both copies of the γ134.5 gene (17).
Cell Infection.
HLF cells were maintained at confluence for 1 week. Glioma cells were maintained in a plateau phase for at least 48 hr. The cells were exposed to 10 plaque-forming units of HSV-1(F) or R3616 for 2 hr. The inoculum then was replaced with the growth DMEM supplemented with heated 10% FCS for 2 hr and reincubated at 37°C for 3 additional hr.
Preparation of RNA.
Conditioned media were discarded from culture flasks, and adherent cells were directly lysed in flasks by TRIzol reagent (GIBCO/BRL) according to the manufacturer’s instructions. After one cycle of deproteinization, RNA was pelleted by isopropanol, rinsed by ethanol, and redissolved in 200 μl of RNAsecure resuspension solution (Ambion, Austin, TX). The RNA preparations were digested with 0.1 units/μl of DNase I, in DNase 1 buffer from CLONTECH. The RNAs were additionally deproteinized in phenol-chloroform-isoamyl alcohol mixture, RNA-pelleted by ethanol, and dissolved in 50–100 μl of RNAsecure™ resuspension solution. RNA concentration was measured spectrophotometrically, assuming that 1A260 is equal to a concentration of 35 μg/ml. Samples were adjusted to a final concentration of 1 mg/ml and stored at −80°C without detectable signs of degradation. All RNA preparations were assessed for integrity by agarose electrophoresis in the presence of RNA standards (GIBCO/BRL). All preparations described in this paper contained intact RNA with distinct 28S, 18S, and 5S bands (data not shown). No detectable differences in electrophoretically defined RNA pattern between noninfected and infected cells were noted.
Preparation of Radiolabeled cDNA Probes and Hybridization with DNA Arrays.
cDNA synthesis was done with Moloney murine leukemia virus reverse transcriptase (SuperScript II, GIBCO/BRL), in the presence of gene-specific primers (CLONTECH) and [32P]-dATP, according to the manufacturer’s protocols. For cDNA synthesis we used equal amounts (5 μg) of total RNA from each sample. cDNA was purified on ChromaSpin-200 diethyl pyrocarbonate-H2O columns (CLONTECH), and fractions corresponding to cDNA were pooled and counted in a Beckman LS 3801 liquid beta-spectrometer. All samples were equalized by total amounts of counts, loaded on DNA array. Typically, we used 15 × 106 cpm of [32P] per membrane. Prehybridization and hybridization of Atlas Human cDNA expression arrays (CLONTECH) were done at 68°C for 1 hr and 16 hr, respectively. Washing of membranes was performed according to the manufacturer’s protocols, except that washing in 0.1% SSC, 0.5% SDS was done twice, for 1 hr each time. DNA arrays were exposed to Kodak Bio Max MS films with Kodak HE intensifying screens at −80°. At least three films were developed from each membrane, with exposure times of 16, 48, and 72 hr. Films were scanned at 600 dpi by using a Mirage II scanner (Umax, Fremont, CA). Data analysis was assisted by use of the AtlasInfo database (http://atlasinfo.clontech.com).
Results
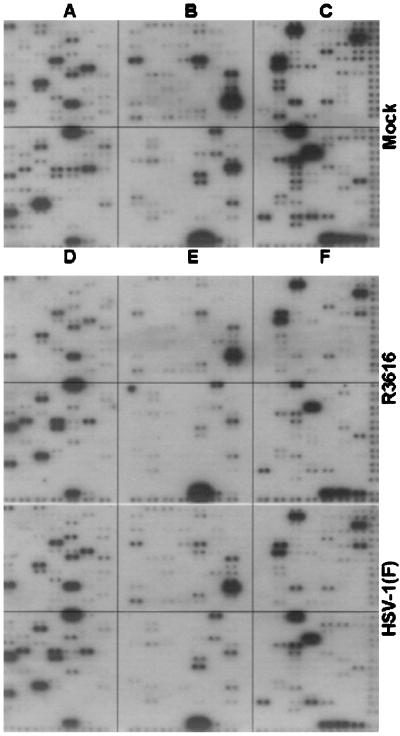
Fig. 1 shows the autoradiograms of the hybridization of labeled cDNAs of HLF mock-infected or infected with HSV-1(F) or R3616. Long exposure of the membranes indicated that we were able to detect signals from approximately 40% of the 588 genes represented in the DNA arrays. The results of hybridizations of cDNAs obtained from the other two cell lines, either mock-infected or infected with HSV-1(F) or with R3616 (data not shown) were, with few exceptions illustrated below, comparable to those shown in Fig. 1. The salient features of the results of these studies were as follows:
Figure 1.
Autoradiographic images of DNA arrays probed with cDNA obtained from HLF mock-infected (Top), infected with the recombinant virus R3616 (Middle), or parent, wild-type virus HSV-1(F) (Bottom). Genes probes are grouped into quadrants as follows: (A) control of cell cycle; (B) signal transduction; (C) apoptosis and DNA repair; (D) transcriptional factors; (E) receptors and cell adhesion; and (F) cytokines and hormones.
(i) As could be expected, there was an overall decrease in the total amount of cell-specific RNA in infected cells as compared with those of mock-infected cells. With few exceptions noted below, we did not observe a major difference in the accumulation of RNA in cells infected with HSV-1(F) as compared with those of the recombinant virus R3616.
(ii) The 588 human genes represented in the microarrays fall into six groups arranged in quadrants and comprising genes whose products are involved in the control of cell cycle and tumor suppression (quadrant A), signal transduction, ion transport, and stress response (quadrant B), apoptosis and DNA repair/recombination (quadrant C), transcriptional factors (quadrant D), receptors, cell-surface antigens, and cell adhesion (quadrant E), and cytokines and hormones (quadrant F). Examination of patterns of hybridization of the cDNAs of all three cell lines indicated that accumulation of cell-specific transcripts at a level significantly above that of mock-infected cells occurred largely in quadrant D and to a much lesser extent in quadrant C.
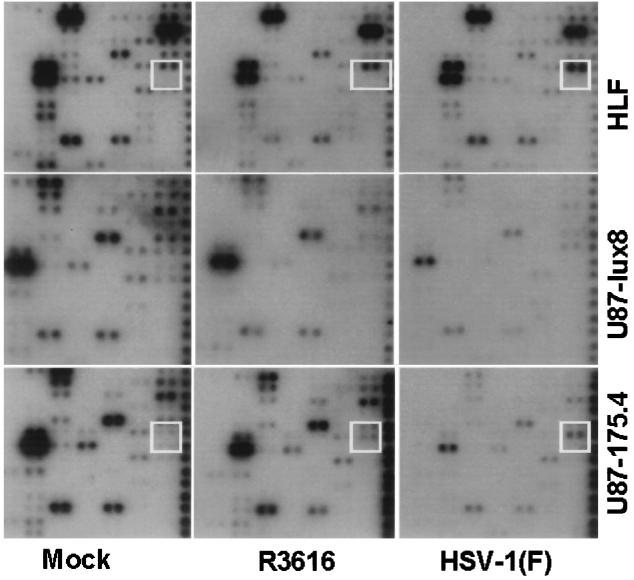
(iii) As noted above, quadrant C contained probes for transcripts of genes involved in apoptosis, stress responses, and DNA repair. The sole transcript accumulating in larger amounts in HLF infected with wild-type virus as compared with those of mock-infected cells was that of GADD45 (Fig. 2, upper two dots in white square). The GADD45 transcript accumulated in decreased amounts in cells infected with the R3616 mutant. This transcript accumulated at higher levels in U87–175.4 cells infected with HSV-1(F) but not in U87-lux8 cells infected with HSV-1(F) or R3616. It is also noteworthy, as discussed later in the text, that there was no increase in the accumulation of GADD153 transcripts (Fig. 2, lower two dots in white square).
Figure 2.
Autoradiographic images from quadrant C containing probes representing genes involved in apoptosis, stress, and DNA repair probed with cDNA obtained from HLF, U87-lux8, and U87–175.4 cells mock-infected or infected with R3616 or HSV-1(F) for 5 hr. The white box surrounds the location of GADD45 (upper two dots) and GADD153 (lower two dots).
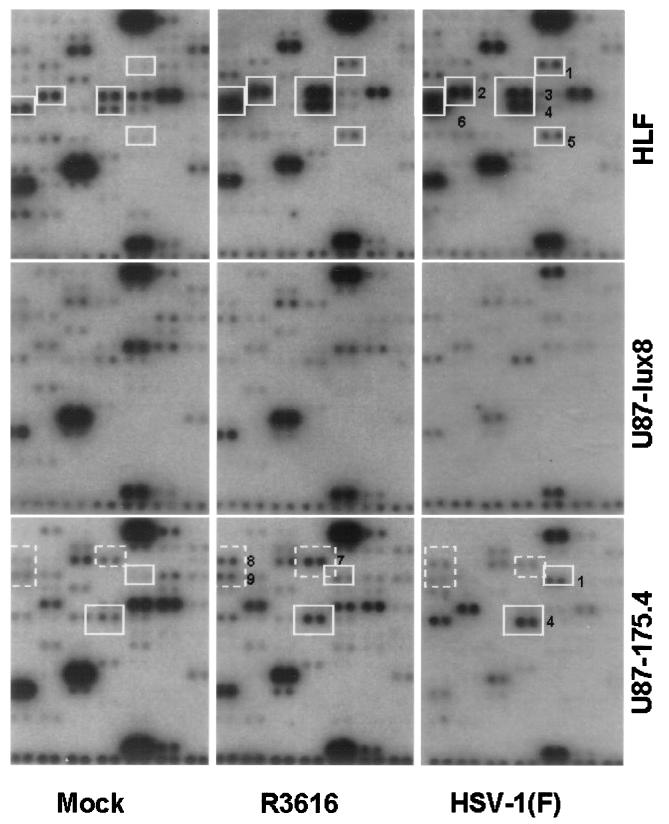
(iv) Quadrant D contained probes for transcripts of genes involved in regulatory processes and transcription. Two observations are of potential significance. First, in HLF cells, several transcripts accumulated in larger amounts in both infected cells as compared with mock-infected cells. These were (Fig. 3, delineated by numbered white squares or quadrangles) the transcripts encoding tristetraproline (no. 1), activating transcription factor-4 (ATF-4) (no. 2), Egr1 (no. 3), ETR101 (no. 4), HOX7 (no. 5), and inhibitor of DNA binding 2 (Id-2) (no. 6). No significant elevation in the level of these transcripts was observed in U87-lux8 cells. In U87-l75.4 cells only two of the transcripts noted above accumulated in larger amounts in both HSV-1(F)-infected cells and in cells infected with R3616. These were tristetraprolin (no. 1) and ETR101 (no. 4). The second observation of particular interest is that several transcripts accumulated in larger amounts in U87–175.4 cells infected with R3616 than in cells either mock-infected or infected with wild-type virus (Fig. 3, dashed white quadrangles). These were transcripts encoding AP-2 (no. 7), CCAAT DNA box-binding protein (no. 8), and Id-3 (no. 9).
Figure 3.
Autoradiographic images of quadrant D containing probes representing genes encoding regulatory proteins and transcription factors. The DNA arrays were probed with cDNA obtained from HLF, U87-lux8, and U87–175.4 cells mock-infected or infected with R3616 or HSV-1(F) for 5 hr. mRNA species that accumulated in HSV-1-infected cells are boxed in white and numbered as follows: 1, tristetraprolin; 2, ATF-4; 3, Egr1; 4, ETR101; 5, HOX7; and 6, Id-2. mRNAs that accumulated specifically U87–175.4 cells infected with R3616 in contrast to mock-infected cells or cells infected with HSV-1(F) infection in are marked by dashed boxes boxed and labeled as follows: 7, AP-2; 8, CCAAT DNA box-binding protein; and 9, Id-3.
(v) The DNA arrays were scanned with the aid of the National Institutes of Health image program. The data were normalized as follows: The ratios of HSV-1(F)-infected to uninfected values for three housekeeping gene transcripts (ribosomal protein S9, Mr 23,000 basic protein, and β-actin) were calculated and averaged. The ratios of HSV-1(F) to uninfected values for individual transcripts boxed in Figs. 2 and 3 were calculated and normalized with respect to that of the averaged housekeeping genes. On that basis, the lowest accumulation considered significant was that of ATF-4 (2.1) whereas the highest was exhibited by Hox-7 (12.1). The normalized value for GADD45 was 4.2. The values were lower for cells infected with R3616 mutant.
Discussion
The salient features of the results were as follows: Notwithstanding a sharp decrease in the accumulation of cellular RNAs resulting from virus-induced degradation of mRNA and inhibition of splicing of nascent transcripts, there was significant accumulation of a small subset of cellular transcripts. The transcripts encode regulatory proteins and transcriptional factors and one stress-response protein (GADD45). The interesting feature of the results is that in the case of HLF cells, the same set of six transcripts accumulated in larger amounts in cells infected with R3616 and HSV-1(F) than in mock-infected cells. In U87–175.4 cells, only two of the six transcripts accumulated in larger amounts after HSV-1(F) infection than in control cells. In contrast, cells infected with the R3616 recombinant accumulated transcripts of three other genes in larger amounts than in either mock-infected or wild-type infected cells. There was no significant accumulation of transcripts above those of mock-infected cell in U87-lux8 cells expressing the wild-type p53 protein. In experiments, however, the accumulation of trnascripts in U87 parental cell line was similar (but not identical) to those reported in this study for HLF described above (data not shown). The difference may reflect the genotype of the selected clone rather than expression of wild-type p53. Moreover, particularly in the U87-lux8 cells infected with wild-type virus, the overall decrease in the accumulation of transcripts was far greater than in cells infected with R3616 recombinant (see Figs. 2 and 3). The conclusions from these observations are that (i) a small number of transcripts accumulate in large amounts notwithstanding the overall shutoff of host RNA accumulation and (ii) the nature and amount of accumulated transcript varies depending on the genotype of the virus and the infected cells.
The nature of the transcripts accumulating in infected cells at levels above those of uninfected cells constitutes a narrow, but useful, window into the interaction of the virus and its host. Specifically:
(i) Id-2, a member of the helix–loop–helix family of transcription factors, forms heterodimers to antagonize other transcriptional factors and acts as an antagonist of multiple tumor suppressor proteins (20, 21). Id-2 binds to the retinoblastoma protein (pRb) and abolishes its growth-suppressing activity. Id2 also binds in vitro pRb-related proteins p107 and p130 (22, 23). Constitutive expression results in decreased levels of cyclin D1 and the loss of cyclin D1-cdk4 complexes (23). Of particular interest is the observation that Id-2 is an inhibitor of tissue-specific gene expression. For example, it is expressed during early fetal development and in induced differentiation of the neuroblastoma cell line SMS-KCNR. Our studies do not differentiate between various members of the Id-2 family. It is noteworthy that Id-2b arises from an intronless gene and hence is less subject to suppression than spliced mRNA.
(ii) Egr-1 and the closely related Egr-2, Egr-3, and Egr-4 are members of a subclass of immediate early, inducible transcription factors. Egr-1, a nuclear zinc-finger protein expressed in response to diverse stimuli and a potential regulator of numerous genes (e.g., IL-2, CD44, intercellular adhesion molecule-1, and tumor necrosis factor genes) is involved in growth, development, differentiation, and transformation suppression activity (24). EGR-1 recognizes and binds to the DNA sequence 5′-CGCCCCCGC-3′ (EGR-site) (25). Egr-1 also plays a putative role in regulation of the immune response (26).
(iii) The ETR101 mRNA is induced by 12-O-tetradecanoyl-phorbol 13-acetate and under certain conditions in the presence of cyclohexamide and is inhibited by protein kinase C inhibitors (27). The ETR101 appears to be derepressed by infection. Its function in the infected cell is not readily apparent.
(iv) HOX7 (MSX1) is a member of the Msx family of proteins containing a homeobox (183-bp highly conserved, helix–turn–helix motif) (28). The protein has been implicated to play a key role in the differentiation, development of certain tissues, and cellular proliferation (29).
(v) AP-2α, a member of a family of three known AP-2 genes (α, β, and γ), is a nuclear transcriptional factor that positively and negatively regulates genes involved in development and cellular differentiation (30). It binds to the consensus sequence CCCAGGC as a dimer. AP-2 binding sites are present in genomes of numerous DNA viruses and the product of at least one virus, the major T antigen of simian virus 40 binds AP-2 and prevents it from binding DNA. In the case of hepatitis B virus, the X protein stimulates AP-2-mediated transcription (31). AP-2 is a negative regulator of transactivation by myc (32). AP-2 is involved in the mammalian stress response, and it is thought that AP-2 suppresses apoptosis by protecting proliferating cells from myc-induced apoptotic cell death.
(vi) CBP (CCAAT binding protein) also known as NEYA, HAP2, etc. as its name indicates, binds a motif present in promoter domain of many unrelated genes (33). It is the β subunit of a CCAAT heterotrimeric protein binding complex.
(vii) ATF-4, a leucine zipper containing a member of ATF/CREB (cAMP responsive element binding protein), binds to the consensus sequence GTGACGTACAG and is present in promoters of both viruses and cells (34, 35).
(viii) Tristetraprolin, a zinc finger protein homologous to ZFP-36 in mouse, is a member of the Cys-Cys-Cys-His (CCCH) zinc finger proteins reported to affect the switch from G0 to G1 (36). Tristetraproline has been reported to negatively regulate tumor necrosis factor α (TNF-α) production by binding the AU-rich element within the 3′ noncoding sequences of TNF-α mRNA (37).
(ix) The acronym GADD stands from growth arrest and damage. All five known GADD genes are induced as a consequence of DNA damage induced by UV light irradiation but only GADD45 is induced by ionizing radiation (38, 39). The observation that GADD45 but not GADD153 is induced during infection of HLF suggests a similarity in the nature of DNA damage induced by infection and ionizing radiation that leads to induction of GADD45 but not of GADD153. Relevant to this report is the observation that GADD45 induction is associated with the activation of the c-jun N-terminal kinase (JNK)/p38 mitogen-activated protein kinase pathway (40, 41). This pathway can result in JNK-mediated apoptosis. GADD45 can be activated by either p53 or transforming growth factor type β.
(x) Id-3, also known as HLH 462, is an immediate early cellular protein (42). Like Id-2 it can form dimers with basic helix–loop–helix proteins to inhibit transcription. Cdk2/cyclin A or E complexes can phosphorylate Id-3. The state of phosphorylation of Id-3 regulates its specificity for inhibiting various basic helix–loop–helix molecules (43).
The studies carried out at this point do not address the question of whether the expression of cellular genes whose transcripts accumulate in infected cells is caused by viral or cellular factors. Many of the transcriptional factors whose transcripts accumulate in infected cells regulate gene expression both negatively and positively are of potential benefit to both host and virus. For example, induction of Id-2 or of Id-3 could benefit both the virus and the cell. The cell would benefit from the shutoff of transcription of viral genes because transcription, ultimately, depends on cellular factors. The benefit to the virus would accrue from Id-2-induced changes in transcriptional program of differentiated cells that could enrich the infected cells with precursors necessary for viral nucleic acid synthesis.
The accumulation of Egr1 may be a consequence of GADD45 induction, because GADD45 can up-regulate expression of Egr1 (44). GADD45 also functions at the G2/M checkpoint to halt cell cycle progression (45). Induction of Egr-1 could benefit the human host through induction of apoptosis in infected cells by transactivation of p53. Activation of Egr-1 also could benefit the virus through induction of key gene encoding proteins useful for viral replication. Even in the case of cellular proteins whose functions are of no apparent benefit to the virus could in fact be co-opted to serve the needs of the invading virus as has been illustrated in the case of protein phosphatase 1 (13, 14). The only possible exception may be GADD45 whose induction may reflect breaks in cellular DNA and an unsuccessful attempt to induce apoptosis. Further studies on the cellular proteins whose transcripts accumulate in infected cells may clarify their role in infected cells.
Acknowledgments
U87-lux8 and U87–175.4 cell lines were kindly provided by Mark Israel (Preuss Laboratory for Molecular Neuro-oncology, University of California, San Francisco). These studies were aided by grants to R.R.W. from the National Cancer Institute (CA71933) and to B.R. from the National Cancer Institute (CA47451, CA71933, and CA78766), and the United States Public Health Service.
Abbreviations
- HSV-1
herpes simplex virus 1
- HLF
human lung fibroblasts
- ATF
activating transcription factor
- ICP
infected cell protein
- Id
inhibitor of DNA binding
References
- 1.Roizman B, Sears A E. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Philadelphia: Lippincott; 1996. pp. 2231–2295. [Google Scholar]
- 2.Read G S, Frenkel N. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schek N, Bachenheimer S L. J Virol. 1985;55:601–610. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong A D, Frenkel N. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strom T, Frenkel N. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwong A D, Kruper J A, Frenkel N. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardwicke M A, Sandri-Goldin R M. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardy W R, Sandri-Goldin R M. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 11.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Nature (London) 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 12.Chou J, Chen J J, Gross M, Roizman B. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Gross M, Roizman B. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He B, Gross M, Roizman B. J Biol Chem. 1998;273:20737–20743. doi: 10.1074/jbc.273.33.20737. [DOI] [PubMed] [Google Scholar]
- 15.Galvan V, Roizman B. Proc Natl Acad Sci USA. 1998;95:3931–3936. doi: 10.1073/pnas.95.7.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galvan V, Brandimarti R, Roizman B. J Virol. 1999;73:3219–3226. doi: 10.1128/jvi.73.4.3219-3226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou J, Kern E R, Whitley R J, Roizman B. Science. 1990;250:1262–1265. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 18.Van Sant C, Kawaguchi Y, Roizman B. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas-Kogan D A, Yount G, Haas M, Levi D, Kogan S S, Hu L, Vidair C, Deen D F, Dewey W C, Israel M A. Int J Radiat Oncol Biol Phys. 1996;36:95–103. doi: 10.1016/s0360-3016(96)00244-1. [DOI] [PubMed] [Google Scholar]
- 20.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 21.Sun X H, Copeland N G, Jenkins N A, Baltimore D. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 23.Lasorella A, Iavarone A, Israel M A. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Calogero A, Ragona G, Adamson E, Mercola D. Crit Rev Oncog. 1996;7:101–125. [PubMed] [Google Scholar]
- 25.Cao X, Mahendran R, Guy G R, Tan Y H. J Biol Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 26.McMahon S B, Monroe J G. J Leukocyte Biol. 1996;60:159–166. doi: 10.1002/jlb.60.2.159. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu N, Ohta M, Fujiwara C, Sagara J, Mochizuki N, Oda T, Utiyama H. J Biol Chem. 1991;266:12157–12161. [PubMed] [Google Scholar]
- 28.Hewitt J E, Clark L N, Ivens A, Williamson R. Genomics. 1991;11:670–678. doi: 10.1016/0888-7543(91)90074-o. [DOI] [PubMed] [Google Scholar]
- 29.Vastardis H, Karimbux N, Guthua S W, Seidman J G, Seidman C E. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P J, Wang C, Tjian R. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 31.Seto E, Mitchell P J, Yen T S B. Nature (London) 1990;344:72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- 32.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mantovani R. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y S, Green M R. Proc Natl Acad Sci USA. 1988;85:3396–3400. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee K A W, Hai T Y, SivaRaman L, Thimmappaya B, Hurst H C, Jones N C, Green M R. Proc Natl Acad Sci USA. 1987;84:8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heximer S P, Forsdyke D R. DNA Cell Biol. 1993;12:73–88. doi: 10.1089/dna.1993.12.73. [DOI] [PubMed] [Google Scholar]
- 37.Carballo E, Lai W S, Blackshear P J. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 38.Fornace A J, Jr, Nebert D W, Hollander M C, Luethy J D, Papathanasiou M, Fargnoli J, Holbrook N J. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papathanasiou M A, Kerr N C, Robbins J H, McBride O W, Alamo I J, Barrett S F, Hickson I D, Fornace A J., Jr Mol Cell Biol. 1991;11:1009–1016. doi: 10.1128/mcb.11.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harkin D P, Bean J M, Miklos D, Song Y H, Truong V B, Englert C, Christians F C, Ellisen L W, Maheswaran S, Oliner J D, Haber D A. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 41.Takekawa M, Saito H. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 42.Christy B A, Sanders L K, Lau L F, Copeland N G, Jenkins N A, Nathans D. Proc Natl Acad Sci USA. 1991;88:1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deed R W, Hara E, Atherton G T, Peters G, Norton J D. Mol Cell Biol. 1997;17:6815–6821. doi: 10.1128/mcb.17.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim C P, Jain N, Cao X. Oncogene. 1998;16:2915–2926. doi: 10.1038/sj.onc.1201834. [DOI] [PubMed] [Google Scholar]
- 45.Wang X W, Zhan Q, Coursen J D, Khan M A, Kontny U, Yu L, Hollander M C, O’Connor P M, Fornace A J, Jr, Harris C C. Proc Natl Acad Sci USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]