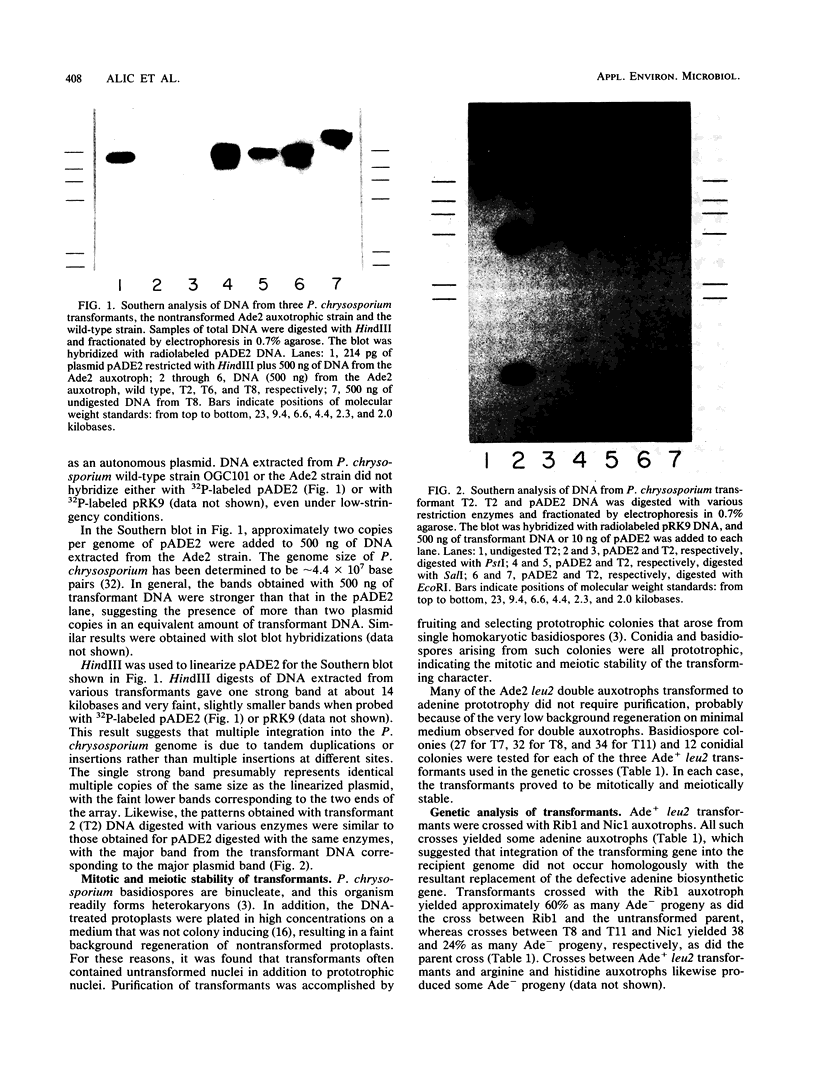
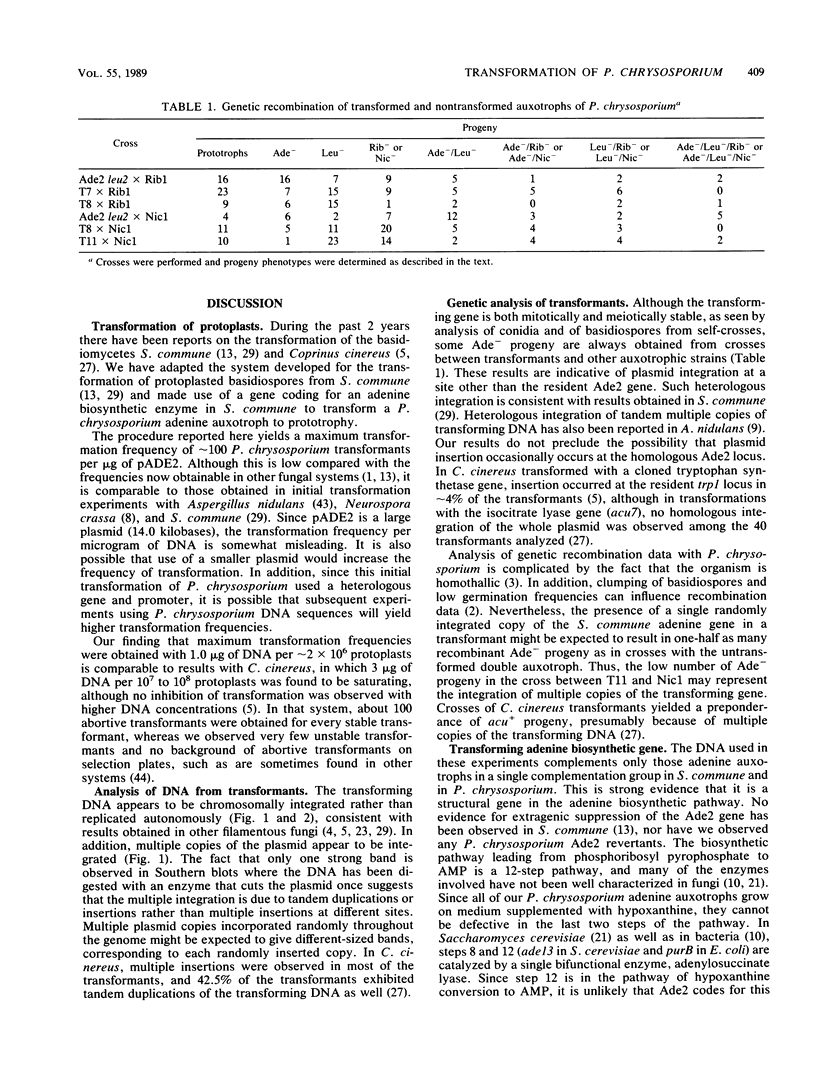
Abstract
Swollen basidiospores of an adenine auxotroph of Phanerochaete chrysosporium were protoplasted with Novozyme 234 and transformed to prototrophy by using a plasmid containing the gene for an adenine biosynthetic enzyme from Schizophyllum commune. Transformation frequencies of 100 transformants per μg of DNA were obtained. Southern blot analysis of DNA extracted from transformants demonstrated that plasmid DNA was integrated into the chromosomal DNA in multiple tandem copies. Analysis of conidia and basidiospores from transformants demonstrated that the transforming character was mitotically and meiotically stable on both selective and nonselective media. Genetic crosses between double mutants transformed for adenine prototrophy and other auxotrophic strains yielded Ade− progeny, which indicated that integration occurred at a site(s) other than the resident adenine biosynthetic gene.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. General method for cloning Neurospora crassa nuclear genes by complementation of mutants. Mol Cell Biol. 1985 Sep;5(9):2272–2278. doi: 10.1128/mcb.5.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic M., Gold M. H. Genetic Recombination in the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1985 Jul;50(1):27–30. doi: 10.1128/aem.50.1.27-30.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic M., Letzring C., Gold M. H. Mating System and Basidiospore Formation in the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1987 Jul;53(7):1464–1469. doi: 10.1128/aem.53.7.1464-1469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance D. J., Turner G. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene. 1985;36(3):321–331. doi: 10.1016/0378-1119(85)90187-8. [DOI] [PubMed] [Google Scholar]
- Binninger D. M., Skrzynia C., Pukkila P. J., Casselton L. A. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 1987 Apr;6(4):835–840. doi: 10.1002/j.1460-2075.1987.tb04828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrens P., Green P. M., Arst H. N., Jr, Scazzocchio C. Heterologous insertion of transforming DNA and generation of new deletions associated with transformation in Aspergillus nidulans. Mol Gen Genet. 1986 Jun;203(3):544–549. doi: 10.1007/BF00422084. [DOI] [PubMed] [Google Scholar]
- Ebbole D. J., Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987 Jun 15;262(17):8274–8287. [PubMed] [Google Scholar]
- Eriksson K. E. Cellulases of fungi. Basic Life Sci. 1981;18:19–32. doi: 10.1007/978-1-4684-3980-9_3. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Froeliger E. H., Muñoz-Rivas A. M., Specht C. A., Ullrich R. C., Novotny C. P. The isolation of specific genes from the basidiomycete Schizophyllum commune. Curr Genet. 1987;12(7):547–554. doi: 10.1007/BF00419565. [DOI] [PubMed] [Google Scholar]
- Glenn J. K., Akileswaran L., Gold M. H. Mn(II) oxidation is the principal function of the extracellular Mn-peroxidase from Phanerochaete chrysosporium. Arch Biochem Biophys. 1986 Dec;251(2):688–696. doi: 10.1016/0003-9861(86)90378-4. [DOI] [PubMed] [Google Scholar]
- Glenn J. K., Gold M. H. Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys. 1985 Nov 1;242(2):329–341. doi: 10.1016/0003-9861(85)90217-6. [DOI] [PubMed] [Google Scholar]
- Gold M. H., Cheng T. M., Alic M. Formation, Fusion, and Regeneration of Protoplasts from Wild-Type and Auxotrophic Strains of the White Rot Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1983 Jul;46(1):260–263. doi: 10.1128/aem.46.1.260-263.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. H., Cheng T. M. Induction of colonial growth and replica plating of the white rot basidiomycete Phanaerochaete chrysosporium. Appl Environ Microbiol. 1978 Jun;35(6):1223–1225. doi: 10.1128/aem.35.6.1223-1225.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. H., Cheng T. M., Mayfield M. B. Isolation and Complementation Studies of Auxotrophic Mutants of the Lignin-Degrading Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1982 Oct;44(4):996–1000. doi: 10.1128/aem.44.4.996-1000.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M. H., Kuwahara M., Chiu A. A., Glenn J. K. Purification and characterization of an extracellular H2O2-requiring diarylpropane oxygenase from the white rot basidiomycete, Phanerochaete chrysosporium. Arch Biochem Biophys. 1984 Nov 1;234(2):353–362. doi: 10.1016/0003-9861(84)90280-7. [DOI] [PubMed] [Google Scholar]
- Kersten P. J., Tien M., Kalyanaraman B., Kirk T. K. The ligninase of Phanerochaete chrysosporium generates cation radicals from methoxybenzenes. J Biol Chem. 1985 Mar 10;260(5):2609–2612. [PubMed] [Google Scholar]
- Kinsey J. A., Rambosek J. A. Transformation of Neurospora crassa with the cloned am (glutamate dehydrogenase) gene. Mol Cell Biol. 1984 Jan;4(1):117–122. doi: 10.1128/mcb.4.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk T. K., Farrell R. L. Enzymatic "combustion": the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- Molskness T. A., Alic M., Gold M. H. Characterization of Leucine Auxotrophs of the White Rot Basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1986 Jun;51(6):1170–1173. doi: 10.1128/aem.51.6.1170-1173.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Rivas A., Specht C. A., Drummond B. J., Froeliger E., Novotny C. P., Ullrich R. C. Transformation of the basidiomycete, Schizophyllum commune. Mol Gen Genet. 1986 Oct;205(1):103–106. doi: 10.1007/BF02428038. [DOI] [PubMed] [Google Scholar]
- Muñoz-Rivas A. M., Specht C. A., Ullrich R. C., Novotny C. P. Isolation of the DNA sequence coding indole-3-glycerol phosphate synthetase and phosphoribosylanthranilate isomerase of Schizophyllum commune. Curr Genet. 1986;10(12):909–913. doi: 10.1007/BF00398288. [DOI] [PubMed] [Google Scholar]
- Paszczyński A., Huynh V. B., Crawford R. Comparison of ligninase-I and peroxidase-M2 from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys. 1986 Feb 1;244(2):750–765. doi: 10.1016/0003-9861(86)90644-2. [DOI] [PubMed] [Google Scholar]
- Raeder U., Broda P. Meiotic segregation analysis of restriction site polymorphisms allows rapid genetic mapping. EMBO J. 1986 Jun;5(6):1125–1127. doi: 10.1002/j.1460-2075.1986.tb04336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. M., Eaton N. R. Functional blocks of the ad-1 and ad-2 mutants of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1969 Feb 7;34(3):301–305. doi: 10.1016/0006-291x(69)90831-6. [DOI] [PubMed] [Google Scholar]
- Smith M. H., Gold M. H. Phanerochaete chrysosporium beta-Glucosidases: Induction, Cellular Localization, and Physical Characterization. Appl Environ Microbiol. 1979 May;37(5):938–942. doi: 10.1128/aem.37.5.938-942.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. L., Schalch H., Gaskell J., Covert S., Cullen D. Nucleotide sequence of a ligninase gene from Phanerochaete chrysosporium. Nucleic Acids Res. 1988 Feb 11;16(3):1219–1219. doi: 10.1093/nar/16.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tien M., Kirk T. K. Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H(2)O(2)-requiring oxygenase. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2280–2284. doi: 10.1073/pnas.81.8.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien M., Tu C. P. Cloning and sequencing of a cDNA for a ligninase from Phanerochaete chrysosporium. Nature. 1987 Apr 2;326(6112):520–523. doi: 10.1038/326520a0. [DOI] [PubMed] [Google Scholar]
- Tilburn J., Scazzocchio C., Taylor G. G., Zabicky-Zissman J. H., Lockington R. A., Davies R. W. Transformation by integration in Aspergillus nidulans. Gene. 1983 Dec;26(2-3):205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Zhang Y. Z., Collins C., Reddy C. A. Analysis of nucleotide sequences of two ligninase cDNAs from a white-rot filamentous fungus, Phanerochaete chrysosporium. Gene. 1987;60(1):93–102. doi: 10.1016/0378-1119(87)90217-4. [DOI] [PubMed] [Google Scholar]