Abstract
Amino acid supply in brain is regulated by the activity of the large neutral amino acid transporter (LAT) at the brain capillary endothelial cell, which forms the blood–brain barrier (BBB) in vivo. Bovine BBB poly(A)+ RNA was isolated from 2.0 kg of fresh bovine brain and size fractionated on a sucrose density gradient, and a size-fractionated bovine BBB cDNA library in the pSPORT vector was prepared. The full-length cDNA encoding the bovine BBB LAT was isolated from this library, and the predicted amino acid sequence was 89–92% identical to the LAT1 isoform. The bovine BBB LAT1 mRNA produced a 10-fold enhancement in tryptophan transport into frog oocytes coinjected with bovine BBB LAT1 mRNA and the mRNA for 4F2hc, which encodes the heavy chain of the heterodimer. Tryptophan transport into the mRNA-injected oocytes was sodium independent and was specifically inhibited by other large neutral amino acids, and the Km of tryptophan transport was 31.5 ± 5.5 μM. Northern blotting with the bovine BBB LAT1 cDNA showed that the LAT1 mRNA is 100-fold higher in isolated bovine brain capillaries compared with C6 rat glioma cells or rat brain, and the LAT1 mRNA was not detected in rat liver, heart, lung, or kidney. These studies show that the LAT1 transcript is selectively expressed at the BBB compared with other tissues, and the abundance of the LAT1 mRNA at the BBB is manyfold higher than that of transcripts such as the 4F2hc antigen, actin, or the Glut1 glucose transporter.
Keywords: biological transport, endothelium, gene expression
Amino acid availability in brain plays an important role in the regulation of several pathways of brain amino acid metabolism, including neurotransmitter synthesis, S-adenosylmethionine production, and protein synthesis (1). The transport of essential amino acids from blood to brain intracellular space involves movement of amino acids through two biological membranes in series: the blood–brain barrier (BBB) and the plasma membrane of brain cells (neurons, glia). The brain capillary endothelial plasma membranes form the BBB in vivo. Because the surface area of the brain cell membrane is orders of magnitude greater than the surface area of the BBB (2), transport across the BBB is the rate-limiting step in amino acid movement from blood to brain intracellular spaces.
The transport of large neutral amino acids across the BBB is mediated by a large neutral amino acid transporter (1), analogous to the leucine (L)-preferring system in peripheral tissues, and now designated LAT for large neutral amino acid transporter (3). However, the L-system at the BBB has a much higher affinity (lower Km) for amino acids as compared with L-systems in peripheral tissues (1). Whereas the Km of the L-system in peripheral tissues is in the 1–10 mM range, the Km of large neutral amino acid transport by the BBB L-system is on the order of 10–100 μM (4). The selective expression of a low-Km LAT at the BBB underlies the selective vulnerability of the brain to the pathologic effects of hyperaminoacidemias (1).
Kanai and coworkers (5) have recently cloned LAT1 from C6 rat glioma cells by using a frog oocyte expression system involving coinjection of mRNA for the 4F2hc antigen. These results indicate that transport of large neutral amino acids is mediated by a system composed of a heterodimer of the 4F2hc heavy chain and the LAT1 light chain (5), similar to other amino acid transporters (6, 7). The Km of leucine transport into frog oocytes injected with LAT mRNA from C6 rat glioma cells is low, 18.1 ± 3.4 μM (5), a value that approximates the Km of leucine transport across the BBB in vivo (4). This similarity in Km values suggests that LAT1 or a related isoform may mediate the transport of large essential neutral amino acids across the BBB in vivo. Therefore, the present studies describe the molecular cloning, sequencing, expression, and Northern blotting of the BBB LAT, and show that the expression of this gene is approximately 100-fold greater than that in other tissues, including C6 rat glioma cells.
Experimental Procedures
Materials.
The SuperScript cloning system and Escherichia coli DH5α competent cells were obtained from GIBCO/BRL and Life Technologies. [32P]dCTP (3,000 Ci/mmol; 1 Ci = 37 GBq), α-[35S]thio-dATP (1,000 Ci/mmol), [3H]tryptophan (20 Ci/mmol), [14C]sucrose (0.6 Ci/mmol), and GeneScreenPlus were purchased from NEN Life Science Products. Oligodeoxynucleotides were custom synthesized at Biosource International (Camarillo, CA). The pCR2.1 vector was obtained from Invitrogen. The full-length rat 4F2hc cDNA (accession no. AB015433) subcloned in pBluescript II SK− was kindly provided by Y. Kanai (Kyorin University, Japan) (6). The BK-actin plasmid was prepared as previously described (8). C6 rat glioma cells were obtained from the American Type Culture Collection. Female oocyte-positive Xenopus frogs (50–70 g) were purchased from Nasco (Fort Atkinson, WI) and maintained at the University of California Los Angeles Vivarium .
Isolation and Fractionation of Bovine Brain Capillary mRNA.
Size fractionation of bovine brain capillary poly(A)+ mRNA was performed by sucrose gradient ultracentrifugation (9). Fresh bovine brain (2,035 g from 12 brains) was obtained for isolation of brain capillaries (Fig. 1B), and 140 μg of poly(A)+ RNA was isolated from the brain capillaries by a single-step method (10). The mRNA was size-fractionated in a 5–20% sucrose gradient, which was formed in a Beckman L8–70M ultracentrifuge using a SW28 rotor at 35,000 rpm at 4°C for 17 hr (155,000 × g), and 10 different sucrose gradient fractions (1 ml each) of various densities were collected. The RNA from fractions 2–3 was used in the construction of a size-fractionated cDNA library in the pSPORT vector.
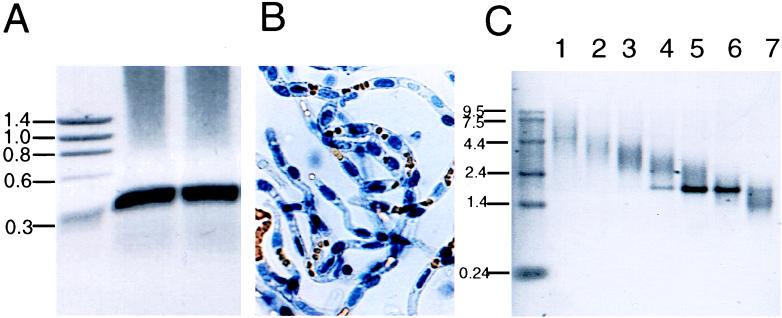
Figure 1.
(A) Rat LAT1 cDNA was generated by reverse transcription–PCR amplification of rat C6 poly(A)+ RNA. Duplicate aliquots of PCR products were analyzed by gel electrophoresis on 0.8% agarose, and the ethidium bromide staining of the gel showed a major ≈290-nt band corresponding to the amplified rat-LAT1 cDNA. The sizes (kb) of the DNA standards are shown on the left. The gel was scanned and the image was inverted. (B) Micrograph of freshly isolated bovine brain capillaries, which are free of adjoining brain tissue; erythrocytes are seen trapped in the capillary lumen. (×100.) Poly(A)+ RNA was isolated from the brain capillary preparation. (C) The brain capillary mRNA was size-fractionated in a 5–20% sucrose gradient, and 0.5-μg aliquots of the first 7 gradient fractions were subjected to agarose gel electrophoresis followed by ethidium bromide staining; RNA standards are shown on the left.
Construction of BBB-cDNA Expression Libraries.
cDNA expression libraries were prepared in the vector pSPORT by using the SuperScript cloning system and sucrose-fractionated BBB-poly(A)+ RNA fractions 2–3 according to the manufacturer’s instructions. Two micrograms of poly(A)+ RNA was used for oligo(dT)-NotI priming, and the reaction was monitored with [32P]dCTP. cDNA fractions containing material >1.6 kb were subcloned in the NotI/SalI cohesive ends of the vector pSPORT to construct a cDNA expression library in E. coli DH5α.
In Vitro Transcription.
Synthetic RNAs (cRNAs) were obtained by in vitro run-off transcription with linearized plasmids (i.e., NotI for bovine LAT subcloned in pSPORT, and XbaI for rat 4F2hc in pBluescript) and T7 RNA polymerase as previously described (8).
Expression of Bovine LAT in Xenopus laevis Oocytes.
Frog oocytes were isolated as described previously (11). Oocytes were injected with 50 nl of water or cRNA solution (i.e., bovine LAT and/or rat 4F2hc) by using a nanoliter injector (World Precision Instruments, Sarasota, FL), and kept in Barth’s/gentamycin solution for 4–5 days at 18°C to allow for expression of the mRNA within the oocyte. For the transport assay, five healthy oocytes were transferred into a vial containing 10 ml of Hepes buffer (0.1 M NaCl/2 mM KCl/1 mM MgCl2/1 mM CaCl2/10 mM Hepes, pH 7.5) and incubated at 22°C for 30 min. The assay was performed with 1.0 ml of Hepes buffer containing 2 μCi of [3H]tryptophan and 0.08 μCi of [14C]sucrose for 3 min at 22°C. Initial time course studies showed [3H]tryptophan uptake was linear for up to 45 min in control oocytes. The reaction was stopped with 10 ml of ice-cold choline buffer (0.1 M choline/2 mM KCl/1 mM MgCl2/1 mM CaCl2/10 mM Hepes, pH 7.5), followed by 3 additional washes with choline buffer. Oocytes were individually dissolved in 0.5 ml of 1 M NaOH for 30 min at 60°C. The radioactivity was measured in a liquid scintillation counter with windows for dual (3H, 14C) counting. The amino acid volume of distribution (VD) in μl per oocyte was calculated as follows:
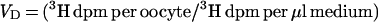 |
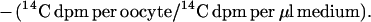 |
The [14C]sucrose provided an internal standard to correct for any incomplete washing. A typical sucrose VD was 0.0079 ± 0.0008 and 0.037 ± 0.008 μl per oocyte at 2 min and 135 min of incubation, respectively (mean ± SE, n = 4). The VD was divided by the incubation time (3 min) to yield the clearance (μl per oocyte per min), and the clearance data were fit to the Michaelis–Menten equation,
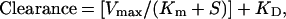 |
where S = the total tryptophan concentration, Vmax is the maximum transport rate, Km is the half-saturation constant, and KD is the constant of nonsaturable transport (1, 4). Data were fit to the equation by using a nonlinear regression analysis (P3R from the BMDP Statistical Software developed by the University of California Los Angeles BMDP Computing Facility). The data were weighted by 1/(clearance)2.
Northern Blot Analysis.
Poly(A)+ RNA was isolated as described previously (10). Northern blots were probed with 32P-labeled bovine LAT, rat 4F2hc, or actin cDNA probes as described previously (10). The x-ray film was scanned and the PhotoShop image was quantified with National Institutes of Health image.
Generation of a Rat LAT1 cDNA Fragment.
A rat LAT1 cDNA was generated by reverse transcription–PCR amplification of rat C6 poly(A)+ RNA as previously described (12, 13). PCR primers were designed to amplify a 291-nt cDNA fragment of the rat LAT1 ORF (nucleotides 1012–1303, GenBank accession no. AB015432). Both forward (5′-GCTGTGGATTTTGGGAACTACC-3′) and reverse (5′-CCACACACAGCCAGTTGAAGAA-3′) primers had a predicted Tm of 66°C, and they possess neither stable stem-loops nor self-complementarity structure. Poly(A)+ RNA was isolated from C6 rat glioma cells as described above. Single-stranded cDNA was synthesized from C6 RNA and PCR amplified with rat LAT1 primers by using protocols previously described (12, 13). Amplification was performed with 35 cycles of denaturation (1 min at 94°C), annealing (2 min at 56°C), and extension (2 min at 72°C). PCR products were analyzed by gel electrophoresis on 0.8% agarose, and a major ≈300-nt band corresponding to the amplified rat LAT1 cDNA was obtained (Fig. 1A). This DNA fragment, named LAT1-6, was isolated from the agarose gel by centrifugation using a Spin X filter unit as previously described (8), and it was directly subcloned into pCR2.1 vector for amplification. The ≈290-nt LAT1-6 insert was released from the plasmid by digestion with EcoRI, a site located at both flanking regions of the single PCR insertion site in pCR2.1, and used for DNA 32P-labeling as described above.
Isolation of Bovine LAT cDNA.
The 32P-labeled clone LAT1-6 was used to screen ≈2.2 × 105 recombinants of a bovine brain capillary λgt11 cDNA library previously constructed and characterized (14). The screening was performed as previously described (14). A positive clone named LAT IV-5 was purified and confirmed in a secondary screening. The positive phage plaque was amplified, and the λgt11 DNA was purified with a λ-isolation kit (Qiagen, Santa Clarita, CA). The λ-DNA insert was released with EcoRI, purified from a 1% agarose gel, and subcloned at the EcoRI site of Bluescript M13+ for DNA sequencing. The LAT IV-5 clone encompasses nucleotides 838-2244 of bovine LAT, and it was used for the isolation of full-length bovine BBB LAT clones from a second bovine brain capillary cDNA library constructed with size-fractionated RNA in the pSPORT vector. Approximately 2.4 × 105 recombinants of the pSPORT cDNA library were screened with the 32P-labeled LAT IV-5 clone by using a colony hybridization technique previously described (14); 186 clones were positive for bovine LAT, and 20 of them were isolated for further characterization. The cDNA inserts were released by double digestion with NotI and SalI and were characterized by ethidium bromide staining of samples separated by agarose gel electrophoresis; the full-length inserts had the same size as the vector and were released with NotI, SalI, and EcoRV triple digestion. Southern blot analysis was performed with 32P-labeled bovine LAT IV-5 insert cDNA under high-stringency conditions as previously described (14).
DNA Sequencing.
DNA sequencing of isolated clones was performed in both directions at the Keck Biotech Resource Lab. (DNA Sequencing Core Facility, Yale University, New Haven, CT), and confirmed by manual DNA sequencing as described previously (14). Manual sequencing was performed with the Sanger method (15) using the Sequenase 2.0 kit and α-[35S]thio-dATP (14). Initial DNA sequencing of was performed with M13 forward and reverse primers. These primers are located in pSPORT at the 5′ and 3′ flanking regions of NotI and SalI sites, respectively. cDNAs were entirely sequenced in both directions by primer walking with custom-synthesized oligodeoxynucleotides as previously described (14); 17- to 20-mers were designed with the program oligos 4.0 so that the primers had a Tm > 60°C in the absence of either stable stem-loops or secondary structures. Similarities with other transporters in GenBank were investigated by using the blast program (National Center for Biotechnology Information), and the phylogenetic analysis of the LAT sequence was performed with the lalig program available through the fasta package at the University of Virginia (16).
Results
A 290-bp DNA fragment was generated by PCR using primers specific for rat LAT1 and cDNA generated from poly(A)+ mRNA isolated from C6 rat glioma cells (Fig. 1A). The 290-bp LAT1 DNA fragment was used to screen a bovine brain capillary cDNA library in the λgt11 vector. Positive clones were screened by autoradiography, and one clone contained the largest insert, 2.0 kb (designated clone LAT IV-5); this insert was subcloned in the Bluescript plasmid. DNA sequencing showed that this 2.0-kb clone contained approximately half of the ORF and the 3′-untranslated region (UTR) of the bovine analog of rat LAT1. Further screening of the λgt11 bovine brain capillary cDNA library did not yield clones containing the full-length bovine LAT cDNA. Therefore, a new library was prepared. Bovine brain capillaries were isolated from fresh bovine brain and purified to homogeneity under RNase-free conditions (Fig. 1B). These capillaries were extracted, bovine brain capillary-derived poly(A)+ RNA was isolated, and 140 μg of this mRNA was size fractionated by sucrose density ultracentrifugation (Fig. 1C). Separate libraries were generated from the respective fractions, and screening of the library generated from fraction 2–3 contained an abundant number of bovine BBB LAT clones. A total of 240,000 clones from the pSPORT size fractionated library were screened with the 32P-labeled clone LAT IV-5, and a total of 186 positive clones were identified, a frequency of 0.078%.
A full-length cDNA was isolated from the bovine brain capillary pSPORT library, and the sequencing results of this clone are shown in Fig. 2. The full-length cDNA is 4039 nucleotides in length and consists of a 5′-UTR segment of 93 nucleotides, an ORF of 1515 nucleotides, and a 3′-UTR of 2431 nucleotides with a short poly(A) tail (Fig. 2). The bovine LAT is composed of 505 amino acids and has a predicted molecular mass of 55,072 daltons. blast search of the High-Throughput Genome Sequence database (hgts) localized the bovine LAT sequence to the clone PAC536B24 (accession no. AC007442), which corresponds to the Homo sapiens chromosome 16q24.3 working draft.
Figure 2.
Nucleotide and deduced amino acid sequence of the bovine LAT. Sequences with high conservation in the UTRs among species are underlined. In the 5′-UTR, nucleotides 28–67 of bovine LAT are 70% and 68% similar to human and rat LAT1, respectively. In the 3′-UTR, the underlined region is 88% similar to the rat LAT1. A putative binding site for the adenosine-uridine binding protein (AUUUA) is also underlined.
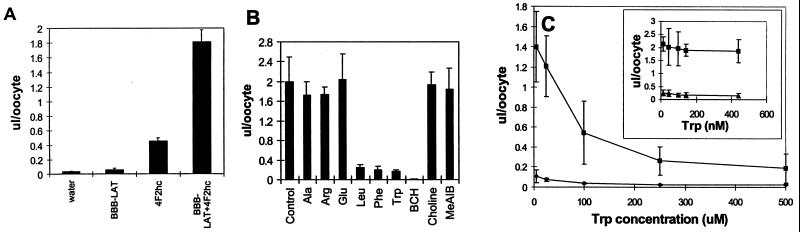
The pSPORT plasmid containing the full-length clone was used to generate by in vitro transcription bovine brain capillary LAT poly(A)+ mRNA, and this mRNA was injected into frog oocytes either alone or with rat 4F2hc mRNA. The uptake of [3H]tryptophan and [14C]sucrose by the mRNA-injected oocytes was measured 4–5 days later. As shown in Fig. 3A, small increases in [3H]tryptophan transport were observed after oocyte injection with either bovine BBB LAT mRNA or rat 4F2hc mRNA. The 4F2hc mRNA alone stimulates transport by forming a dimer with the endogenous frog LAT (5). However, there was a marked increase in [3H]tryptophan transport into the oocyte after the coinjection of 4F2hc and bovine BBB LAT mRNA (Fig. 3A). The [3H]tryptophan transport into the oocytes coinjected with bovine BBB LAT mRNA and 4F2hc mRNA was not inhibited by 500 μM concentrations of small neutral amino acid (alanine), basic amino acid (arginine), or acidic amino acid (glutamate), but was inhibited by large neutral amino acids such as leucine, phenylalanine, tryptophan, or 2-aminobicyclo [2.2.1]heptane-2-carboxylic acid (BCH), a leucine (L)-system-preferring synthetic amino acid (Fig. 3B). The stoichiometric replacement of sodium cations in the transport medium with choline cations did not cause any inhibition of tryptophan transport into the mRNA-injected oocytes. A model amino acid that is specific for the alanine (A)-preferring system, N-methylaminoisobutyric acid (MeAIB), did not compete for tryptophan transport into the mRNA-injected oocytes (Fig. 3B). A tryptophan saturation curve was measured at both nM and μM concentrations of tryptophan for both oocytes injected with 4F2hc alone or oocytes injected with 4F2hc plus bovine BBB LAT mRNA, and these saturation curves are shown in Fig. 3C. Nonlinear regression analysis of these saturation data showed that the Km of tryptophan transport into the oocytes injected with either 4F2hc mRNA alone or 4F2hc mRNA plus bovine BBB LAT mRNA was not significantly different, but there was a 10-fold increase in the Vmax in the oocytes injected with both 4F2hc mRNA and bovine BBB LAT mRNA (Table 1).
Figure 3.
Expression of bovine LAT in Xenopus laevis oocytes. Oocytes were injected with 50 nl water or cRNA solution containing 80 ng of bovine LAT and/or 10 ng of rat 4F2hc and incubated for 4 days at 18°C. (A) The volume of distribution (VD) of [3H]tryptophan in oocytes is increased after the coinjection of 4F2hc mRNA and bovine LAT mRNA. (B) Inhibition of bovine LAT-mediated [3H]tryptophan uptake by amino acids added in 500 μM concentrations. BCH, 2-aminobicyclo[2.2.1]heptane-2-carboxylic acid; MeAIB, N-methylaminoisobutyric acid. (C) Kinetic analysis of the LAT-mediated uptake of [3H]tryptophan. Oocytes were injected with 4F2hc cRNA alone (♦, ▴ in Inset) or in combination with bovine LAT (■). Competition studies were performed by varying the concentration of [3H]tryptophan (14–140 nM) or by increasing the levels of unlabeled tryptophan (0.3–500 μM). The Inset represents the beginning of the curve shown in the main figure, and it is used to demonstrate that there are no changes in Km. Each bar or point represents mean ± SE, n = 5.
Table 1.
Michaelis–Menten parameters of [3H]tryptophan transport into frog oocytes injected with 4F2hc mRNA either alone or with bovine BBB LAT mRNA
| mRNA | Km, μM | Vmax, pmol/oocyte/min | KD, μl/oocyte/min |
|---|---|---|---|
| 4F2hc | 35.9 ± 11.7 | 1.9 ± 0.5 | 0 |
| 4F2hc + LAT | 31.5 ± 5.5 | 19.4 ± 3.3 | 0.024 ± 0.009 |
Results are mean ± SE as determined by nonlinear regression analysis of the saturation data in Fig. 3C.
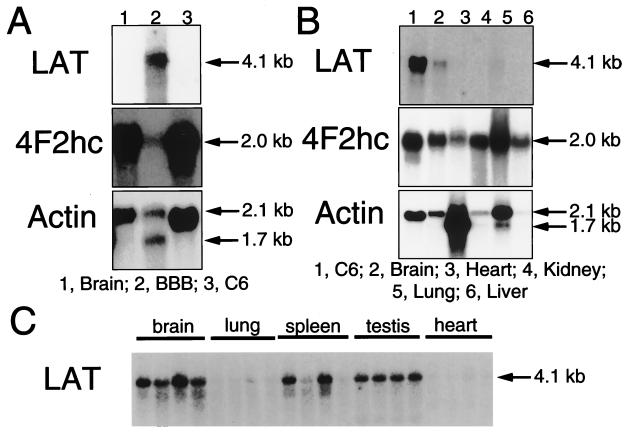
Northern blotting studies with the 32P-labeled cloned LAT IV-5 showed that the mRNA for bovine LAT was profoundly up-regulated in brain capillaries (Fig. 4). The top blot of Fig. 4A shows the 4.1-kb LAT transcript in mRNA obtained from freshly isolated bovine brain capillaries (Fig. 1B), and this transcript was detected after only a 2-hr exposure at room temperature using 2 μg of poly(A)+ mRNA per lane. At this short exposure of the filter to the x-ray film, no detectable LAT mRNA was observed for either total rat brain or C6 rat glioma cells (Fig. 4A, top blot). Conversely, both 4F2hc and actin transcripts were detected in total rat brain and C6 glioma mRNA samples (Fig. 4A, middle and bottom blots). Unexpectedly, the level of 4F2hc mRNA in the brain capillary (BBB) sample was <10% of the value of the 4F2hc mRNA in either total brain or C6 glioma cells, and the BBB 4F2hc transcript signal was detected only after overexposure of the film (Fig. 4A, middle blot). The LAT mRNA was detected in both C6 glioma cells and total rat brain after prolonged exposure of the film as shown in the top blot of Fig. 4B. In these studies, 2 μg of poly(A)+ mRNA was applied per lane and the x-ray film was exposed for 7 days at −70°C. There was no measurable expression of the LAT mRNA at this exposure in rat heart, kidney, liver, or lung (Fig. 4B), although 4F2hc mRNA was detected in all samples and actin transcript was measurable in all samples except in rat liver. A clear LAT mRNA signal was detected in total rat brain when 10 μg of poly(A)+ mRNA was applied per lane and the filter was exposed for 7 days at −70°C as shown in Fig. 4C. The LAT mRNA was comparable in RNA fractions from four different rats, and the LAT mRNA was not detected in lung or heart, but was measurable in rat spleen or testis at the 7- day exposure using 10 μg of poly(A)+ RNA per lane (Fig. 4C); this filter was also used for actin Northern blotting, which showed comparable actin signals in all rat tissues.
Figure 4.
Northern blot analyses of poly(A)+ RNA using 32P-labeled bovine LAT clone IV-5, 4F2hc, or actin. Northern blotting was performed with 2 μg (A and B) or 10 μg (C) of poly(A)+ RNA. Membranes were exposed to Kodak BioMax film and intensifying screens as follows: A, LAT, 2 hr at 22°C; 4F2hc and actin, 20 hr at −70°C; B, LAT, 7 days at −70°C; 4F2hc and actin, 20 hr at −70°C; and C, LAT, 7 days at −70°C. The mRNA was derived from rat brain, lung, spleen, testis, or heart; C6 rat glioma cells; or bovine brain capillaries (BBB).
Discussion
The results of these studies are consistent with the following conclusions. First, there is a profound up-regulation of the LAT mRNA at the BBB compared with any other tissue (Fig. 4). Second, the cloned and expressed bovine BBB LAT is sodium independent, is specific for large neutral amino acids, and has a high affinity with a Km of 31.5 ± 5.5 μM (Fig. 3, Table 1), and this Km value correlates with Km measurements determined for tryptophan transport across the BBB in vivo (4). Third, the amino acid sequence of the bovine LAT corresponds to the LAT1 isoform, and this sequence is strongly conserved across four species (rat, human, mouse, and bovine), and there is also phylogenetic conservation of nucleotide sequence in both the 5′-UTR and the 3′-UTR of the LAT1 mRNA (Fig. 2).
The Northern blotting studies in Fig. 4 show that the LAT1 transcript is selectively expressed at the BBB in vivo. The level of LAT1 transcript in C6 rat glioma cells in tissue culture was as high as any other tissue in the rat (Fig. 4). Rat LAT1 corresponds to TA1 (17), an oncofetal antigen that is expressed primarily in fetal tissues and cancer cells such as C6 glioma cells (5). The LAT1 transcript is also expressed in barrier tissues such as the testis and the placenta (5). The expression of LAT1 in human placenta approximates LAT1 expression in human brain (18), and the expression of the LAT1 mRNA in brain is comparable to the expression in C6 glioma cells (Fig. 4). However, the expression of LAT1 in C6 glioma cells is manyfold less than the expression of the LAT1 mRNA at the BBB, as the time required to develop the BBB LAT1 Northern blot is 1/100 of the time required to develop the LAT1 Northern blot for C6 cells (Fig. 4). The high abundance of the LAT1 transcript at the BBB was also suggested by the high frequency (0.078%) of the LAT1 transcript in the pSPORT library (Results). The ratio of 4F2hc mRNA to LAT1 mRNA is ≫ 1 in C6 glioma cells, but is ≪ 1 at the BBB (Fig. 4). If the relative lack of 4F2hc mRNA correlates with the lack of 4F2hc protein at the BBB, then the availability of 4F2hc protein may be rate limiting in formation of the LAT1 heterodimer.
The selective distribution of the LAT1 transcript at the BBB is also shown by the absence of the LAT1 transcript in liver, heart, lung, or kidney in the rat (Fig. 4). LAT1 expression in brain cells (neurons, glial cells) in vivo may also be negligible, as the brain LAT1 signal seen after a 7-day exposure may represent solely LAT1 mRNA derived from the capillaries in brain (Fig. 4). This issue can be resolved by Northern blotting of capillary-depleted mRNA as described previously (19). These considerations suggest that other unknown LAT1 isoforms function in non-BBB tissues to mediate the cell membrane transport of large neutral amino acids. Recently, the cDNAs for rat and human LAT2 have been cloned (20, 21), and it has been suggested that LAT2 is the LAT isoform that functions at the BBB (20). However, several lines of evidence indicate LAT1 is the BBB LAT isoform. First, LAT1, but not LAT2, is selective for the large neutral amino acids (refs. 5, 20, and 21; Fig. 3B), and this correlates with the greater transport of large neutral amino acids compared with small neutral amino acids at the BBB in vivo (22). Second, the concentration of LAT1 mRNA at the BBB is extraordinarily high relative to any other tissue (Fig. 4). Third, LAT2 is a high-Km system, as the Km for leucine transport by LAT2, 120 ± 34 μM (20), is up to 10-fold greater than the Km of leucine transport by LAT1 (5), and the low Km of the LAT1 isoform correlates with the low Km of amino acid transport through the BBB in vivo (1). The Km of large neutral amino acid transport into peripheral tissues in vivo is 10- to 100-fold greater than the Km of large neutral amino acid transport at the BBB (1). The Km of the cloned and expressed bovine BBB LAT1 for tryptophan, 31.5 ± 5.5 μM (Table 1), approximates the Km of tryptophan transport across the rat BBB in vivo, 48 ± 6 μM, as determined with the carotid artery injection technique (4), since the in vivo value is an overestimate of the actual Km secondary to mixing of the injection solution with rat plasma in vivo (23). The BBB Km for the large neutral amino acids is approximately the existing plasma concentration of the amino acids, which means the BBB LAT1 is normally heavily saturated by the existing concentrations of circulating large neutral amino acids. Conversely, the LAT in peripheral tissue such as liver, heart, lung, or kidney operates far from saturation, since the Km in these tissues is manyfold greater than the existing plasma amino acid concentrations (1). The high affinity (low Km) of large neutral amino acid transport across the BBB provides the physical basis for the selective vulnerability of the brain to the pathologic effects of hyperaminoacidemias (24). Amino acid supply in brain is altered by the increase in plasma concentration of a single large neutral amino acid, as in phenylketonuria, which causes selective competition effects at the BBB relative to peripheral tissues.
The amino acid sequence of the BBB LAT1 is strongly conserved across species, as there is 92% identity of the amino acid sequence in comparing bovine and human LAT1 (18), and an 89% identity in comparing bovine LAT1 with rat (5) or mouse LAT1 (25). The bovine BBB LAT1 transcript has a long 3′-UTR of over 2400 nt (Fig. 2). There is 54% identity over 1448 nt of 3′-UTR in comparing bovine and human LAT1 sequences (18). Similarly, there is a 70% identity over a stretch of 40 nt in the 5′-UTR in comparing the LAT1 transcript in bovine and human (Fig. 2). This finding suggests that the 5′-UTR or 3′-UTR contains cis-regulatory sequences that mediate either the translational efficiency or the stability of the BBB LAT1 transcript. There is a single AUUUA pentameric sequence approximately 185 nt from the poly(A) tail of the transcript (Fig. 2). The AUUUA cis element binds to a series of cellular AU-binding proteins, which participate in the regulation of gene expression at the posttranscriptional level affecting mRNA stability (26, 27).
In summary, these studies describe the cloning and expression of the bovine BBB LAT1 full-length cDNA, and they show that the expression of the LAT1 transcript at the BBB is unexpectedly high, with a >100-fold enrichment at the BBB compared with other tissues such as C6 rat glioma cells. The x-ray film must be exposed 10-fold longer to detect BBB 4F2hc or actin as compared with LAT1 (Fig. 4), which indicates the abundance of the LAT1 transcript at the BBB is much higher than that for either the 4F2hc or actin transcript. The BBB LAT1 mRNA is also manyfold more abundant than the Glut1 transcript at the BBB (19), and Glut1 is the predominant glucose transporter at the BBB in vivo (28). For example, a Glut1 Northern blot of bovine brain capillary poly(A)+ mRNA must be developed several days at −70°C (10), but the LAT1 Northern blot is developed in only 2 hr at room temperature (Fig. 4A). The higher level of LAT1 mRNA, relative to Glut1 mRNA, at the BBB may not be correlated with a higher amount of LAT1 transporter protein, compared with the Glut1 protein, because the maximal transport velocity (Vmax) for d-glucose transport at the BBB is much higher than the Vmax for large neutral amino acid transport (1). However, the abundant LAT1 mRNA at the BBB may mean this transcript has a high turnover rate. The long 3′-UTR of the BBB LAT1 transcript suggests that one mechanism of regulation of LAT1 gene expression at the BBB may be posttrancriptional. The regulation of BBB LAT1 gene expression may play an important role in the adaptive response of the brain to abnormal plasma amino acid supply. This would be essential because the rates of amino acid incorporation into brain proteins by means of cerebral protein synthesis are about the same as the rates of amino acid influx across the BBB in vivo (24). The brain is selectively spared from the hypoaminoacidemia of malnutrition (29), which may be due to a up-regulation of gene expression of the BBB LAT1 mRNA.
Acknowledgments
Daniel Jeong skillfully prepared the manuscript and Margarita Tayag provided expert technical assistance. Dr. Ernest Wright provided valuable discussions. This work was supported by National Institutes of Health Grant NS-25554.
Abbreviations
- BBB
blood–brain barrier
- LAT
large neutral amino acid transporter
- UTR
untranslated region
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF174615).
References
- 1.Pardridge W M. Physiol Rev. 1983;63:1481–1535. doi: 10.1152/physrev.1983.63.4.1481. [DOI] [PubMed] [Google Scholar]
- 2.Lund-Anderson H. Physiol Rev. 1979;59:305–352. doi: 10.1152/physrev.1979.59.2.305. [DOI] [PubMed] [Google Scholar]
- 3.Christensen H N. J Exp Biol. 1994;196:51–57. doi: 10.1242/jeb.196.1.51. [DOI] [PubMed] [Google Scholar]
- 4.Miller L P, Pardridge W M, Braun L D, Oldendorf W H. J Neurochem. 1985;45:1427–1432. doi: 10.1111/j.1471-4159.1985.tb07209.x. [DOI] [PubMed] [Google Scholar]
- 5.Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. J Biol Chem. 1998;273:23629–23632. doi: 10.1074/jbc.273.37.23629. [DOI] [PubMed] [Google Scholar]
- 6.Wells R G, Lee W-S, Kanai Y, Leiden J M, Hediger M A. J Biol Chem. 1992;267:15285–15288. [PubMed] [Google Scholar]
- 7.Malandro M S, Kilberg M S. Annu Rev Biochem. 1996;65:305–336. doi: 10.1146/annurev.bi.65.070196.001513. [DOI] [PubMed] [Google Scholar]
- 8.Boado R J, Tsukamoto H, Pardridge W M. J Neurochem. 1996;67:1335–1343. doi: 10.1046/j.1471-4159.1996.67041335.x. [DOI] [PubMed] [Google Scholar]
- 9.Brakke M K, van Pelt N. Anal Biochem. 1970;38:56–64. doi: 10.1016/0003-2697(70)90155-7. [DOI] [PubMed] [Google Scholar]
- 10.Boado R J, Pardridge W M. J Neurochem. 1991;57:2136–2139. doi: 10.1111/j.1471-4159.1991.tb06433.x. [DOI] [PubMed] [Google Scholar]
- 11.Hediger M A, Coady M J, Ikeda T S, Wright E M. Nature (London) 1987;330:379–381. doi: 10.1038/330379a0. [DOI] [PubMed] [Google Scholar]
- 12.Boado R J, Pardridge W M. J Neurochem. 1994;62:2085–2090. doi: 10.1046/j.1471-4159.1994.62062085.x. [DOI] [PubMed] [Google Scholar]
- 13.Boado R J, Golden P L, Levin N, Pardridge W M. J Neurochem. 1998;71:1761–1764. doi: 10.1046/j.1471-4159.1998.71041761.x. [DOI] [PubMed] [Google Scholar]
- 14.Boado R J, Pardridge W M. Mol Cell Neurosci. 1990;1:224–232. doi: 10.1016/1044-7431(90)90005-o. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklen S, Coulson H R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Miller W. Adv Appl Math. 1991;12:373–381. [Google Scholar]
- 17.Sang J, Lim Y-P, Panzica M, Finch P, Thompson N L. Cancer Res. 1995;55:1152–1159. [PubMed] [Google Scholar]
- 18.Prasad P D, Wang H, Huang W, Kekuda R, Rajan D P, Leibach F H, Ganapathy V. Biochem Biophys Res Commun. 1999;255:283–288. doi: 10.1006/bbrc.1999.0206. [DOI] [PubMed] [Google Scholar]
- 19.Boado R J, Pardridge W M. Biochem Biophys Res Commun. 1990;166:174–179. doi: 10.1016/0006-291x(90)91927-k. [DOI] [PubMed] [Google Scholar]
- 20.Segawa H, Fukasawa Y, Miyamoto K, Takeda E, Endou H, Kanai Y. J Biol Chem. 1999;274:19745–19751. doi: 10.1074/jbc.274.28.19745. [DOI] [PubMed] [Google Scholar]
- 21.Pineda M, Fernandez E, Torrents D, Estevez R, Lopez C, Camps M, Lloberas J, Zorzano A, Palacin M. J Biol Chem. 1999;274:19738–19744. doi: 10.1074/jbc.274.28.19738. [DOI] [PubMed] [Google Scholar]
- 22.Oldendorf W H. Am J Physiol. 1971;221:1629–1639. doi: 10.1152/ajplegacy.1971.221.6.1629. [DOI] [PubMed] [Google Scholar]
- 23.Pardridge W M, Landaw E M, Miller L P, Braun L D, Oldendorf W H. J Cereb Blood Flow Metab. 1985;5:576–583. doi: 10.1038/jcbfm.1985.86. [DOI] [PubMed] [Google Scholar]
- 24.Pardridge W M. Neurochem Res. 1998;23:635–644. doi: 10.1023/a:1022482604276. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura E, Sato M, Yang H, Miyagawa F, Harasaki M, Tomita K, Matsuoka S, Noma A, Iwai K, Minato N. J Biol Chem. 1999;274:3009–3016. doi: 10.1074/jbc.274.5.3009. [DOI] [PubMed] [Google Scholar]
- 26.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 27.Gillis P, Malter J S. J Biol Chem. 1991;266:3172–3177. [PubMed] [Google Scholar]
- 28.Pardridge W M, Boado R J, Farrell C R. J Biol Chem. 1990;265:18035–18040. [PubMed] [Google Scholar]
- 29.Freedman L S, Samuels S, Fish I, Schwartz S, Lange B, Katz M, Morgan L. Science. 1980;207:902–904. doi: 10.1126/science.6766565. [DOI] [PubMed] [Google Scholar]