Abstract
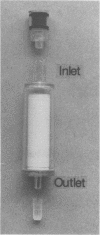
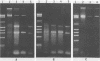
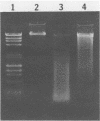
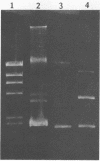
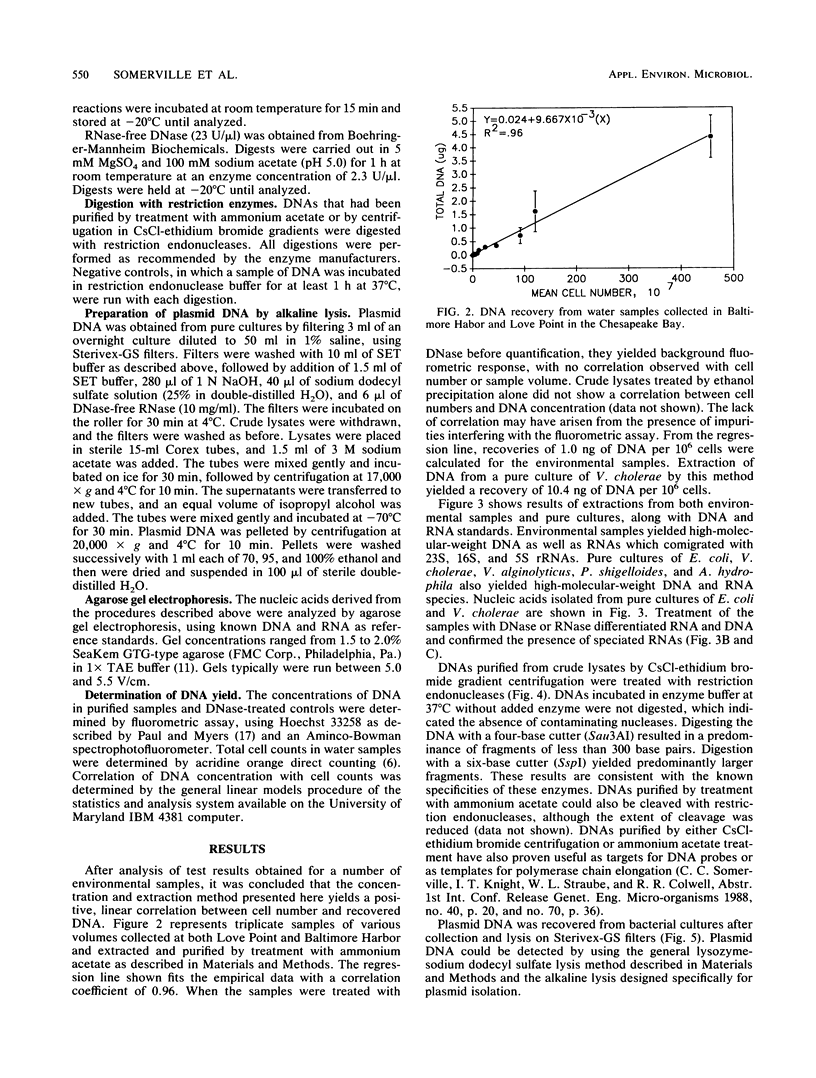
Direct isolation of nucleic acids from the environment may be useful in several respects, including the estimation of total biomass, detection of specific organisms and genes, estimations of species diversity, and cloning applications. We have developed a method that facilitates the concentration of microorganisms from aquatic samples and the extraction of their nucleic acids. Natural water samples of 350 to greater than 1,000 ml are concentrated on a single cylindrical filter membrane (type SVGS01015; Millipore Corp., Bedford, Mass.), and cell lysis and proteolysis are carried out within the filter housing. Crude, high-molecular-weight nucleic acid solutions are then drawn off the filter. These solutions can be immediately analyzed, concentrated, or purified, depending on the intended application. The method is simple, rapid, and economical and provides high-molecular-weight chromosomal DNA, plasmid DNA, and speciated RNAs which comigrate with 5S, 16S, and 23S rRNAs. The methods presented here should prove useful in studying both the ecology and the phylogeny of microbes that resist classical culture methods.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker R. M., Singleton F. L., Hood M. A. Effects of nutrient deprivation on Vibrio cholerae. Appl Environ Microbiol. 1983 Oct;46(4):930–940. doi: 10.1128/aem.46.4.930-940.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson R. L., Buckley E. N., Palumbo A. V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984 Jan;47(1):49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A., Comeau D. E., Hagström A., Chan A. M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988 Jun;54(6):1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra J. A., de Groot R. J., Jacobs L., Kusters J. G., Niesters H. G., van der Zeijst B. A. Synthesis of long cDNA from viral RNA template. Gene Anal Tech. 1988 May-Jun;5(3):57–61. doi: 10.1016/0735-0651(88)90017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., DeFlaun M. F. Dynamics of extracellular DNA in the marine environment. Appl Environ Microbiol. 1987 Jan;53(1):170–179. doi: 10.1128/aem.53.1.170-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Lane D. J., Olsen G. J., Pace N. R. Analysis of hydrothermal vent-associated symbionts by ribosomal RNA sequences. Science. 1984 Apr 27;224(4647):409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Lane D. J., Olsen G. J., Pace N. R. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl Environ Microbiol. 1985 Jun;49(6):1379–1384. doi: 10.1128/aem.49.6.1379-1384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrella F., Morita R. Y. Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl Environ Microbiol. 1981 Feb;41(2):518–527. doi: 10.1128/aem.41.2.518-527.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]