Abstract
Cryptochromes regulate the circadian clock in animals and plants. Humans and mice have two cryptochrome (Cry) genes. A previous study showed that mice lacking the Cry2 gene had reduced sensitivity to acute light induction of the circadian gene mPer1 in the suprachiasmatic nucleus (SCN) and had an intrinsic period 1 hr longer than normal. In this study, Cry1−/− and Cry1−/−Cry2−/− mice were generated and their circadian clocks were analyzed at behavioral and molecular levels. Behaviorally, the Cry1−/− mice had a circadian period 1 hr shorter than wild type and the Cry1−/−Cry2−/− mice were arrhythmic in constant darkness (DD). Biochemically, acute light induction of mPer1 mRNA in the SCN was blunted in Cry1−/− and abolished in Cry1−/−Cry2−/− mice. In contrast, the acute light induction of mPer2 in the SCN was intact in Cry1−/− and Cry1−/−Cry2−/− animals. Importantly, in double mutants, mPer1 expression was constitutively elevated and no rhythmicity was detected in either 12-hr light/12-hr dark or DD, whereas mPer2 expression appeared rhythmic in 12-hr light/12-hr dark, but nonrhythmic in DD with intermediate levels. These results demonstrate that Cry1 and Cry2 are required for the normal expression of circadian behavioral rhythms, as well as circadian rhythms of mPer1 and mPer2 in the SCN. The differential regulation of mPer1 and mPer2 by light in Cry double mutants reveals a surprising complexity in the role of cryptochromes in mammals.
Keywords: gene targeting, photoreceptor, suprachiasmatic nucleus
Circadian rhythms are oscillations with daily periodicities in physiological and behavioral functions of organisms (1–3). The rhythms are generated by a cell-autonomous circadian oscillator (4) that is synchronized with the environment by light. Recently, it was proposed that, in mammals, the nonopsin pigments, cryptochrome blue-light photoreceptors (5, 6), may be the photoactive pigments that synchronized the molecular oscillator and, ultimately, the organismic circadian rhythm with the daily light–dark cycle (7, 8). In humans and mice there are two genes encoding the apoproteins of the cryptochromes: CRY1 and CRY2 in humans and Cry1 and Cry2 in mice (6, 8, 9). Both genes are expressed throughout the body (8–11). Of particular interest, both Cry1 and Cry2 are expressed at high levels in the ganglion cells and the inner nuclear layer of the retina, which are known to be important for circadian photoreception, and Cry1 is expressed with a robust circadian rhythm in the suprachiasmatic nucleus (SCN) (8).
In a previous study (12), we found that mice lacking a functional Cry2 gene (i) had reduced sensitivity to acute light induction of the clock gene mPer1 (mouse period gene 1) in the SCN, (ii) had an intrinsic circadian period about 1 hr longer than normal, and (iii) exhibited high amplitude phase shifts in response to light pulses administered at circadian time (CT) 17. These data, and related findings in cryptochrome mutants of Drosophila melanogaster (13) and Arabidopsis thaliana (14), supported the notion that cryptochromes are major photoreceptors for circadian entrainment in animals and plants. Furthermore, the longer than wild-type circadian period in Cry2−/− mice under constant darkness (DD) and their high-amplitude phase shifts both suggested that cryptochromes could also (or instead) be part of the molecular clock that generates circadian periodicity independently of light (12). That the Cry2−/− mice could be entrained to a 12-hr light/12-hr dark (LD12:12) cycle indicated that the CRY1 protein or other pigments, in addition to the CRY2 protein, participated in photoentrainment. Indeed, in Drosophila, genetic evidence indicates that both the flavin/pterin-based cryptochrome and the retinal-based rhodopsin contribute to circadian photoentrainment (13, 15, 16).
To learn about the roles of cryptochromes in the circadian system, mutant mice lacking CRY1, CRY2, or both CRY1 and CRY2 proteins have been generated and analyzed. Our data show that Cry1−/− mice had circadian periods shorter than wild type and Cry1−/−Cry2−/− mice were arrhythmic under DD. Biochemically, the mPer1 transcript in the SCN exhibited day–night differences but was poorly induced at Zeitgeber time (ZT) 18 in Cry1−/− mice and was chronically elevated and not inducible with acute light pulses in Cry1−/−Cry2−/− mice. In contrast, the mPer2 transcript levels in the SCN exhibited robust day–night differences and light induction in both Cry1−/− and Cry1−/−Cry2−/− mice. Under DD conditions, both mPer1 and mPer2 levels in the SCN were at constant levels in the Cry1−/−Cry2−/− mice; mPer1 was elevated and mPer2 was intermediate. These data provide a molecular explanation for the arrhythmicity in the double mutant we have observed in this study as well as a study that was published (17) while this manuscript was in preparation. Importantly, our data indicate that CRY1 and CRY2 play a central role in the circadian clock system of mammals: they are required for the expression of sustained circadian rhythms and they differentially regulate mPer1 and mPer2 gene expression.
Materials and Methods
Generation of Cry1−/− Mice.
A full-length Cry1−/− cDNA (8) was used to screen a λ-mouse genomic library of the 129/Sv strain. Two phage clones (nos. 29 and 64) carrying genomic fragments overlapping with Cry1 were isolated. However, the two fragments did not overlap; a 13-kb spacing in the mouse genome between the cloned sequences was identified by long PCR. The nucleotide sequence of the ends of the two genomic fragments revealed two exons corresponding to the cDNA sequence of 600–685 and 1646–1715 bp in clones 29 and 64, respectively. Thus, the 13-kb genomic region contains exons corresponding to cDNA sequence 686–1646, which includes the region encoding the FAD-binding domain of the protein (18). We constructed a plasmid (p8529164) that lacked this 13-kb genomic region by subcloning the entire genomic fragment of clone 29 (the 5′ arm) and a 1.5-kb SalI/XhoI fragment of clone 64 (the 3′ arm) into the NotI and XhoI sites, respectively, of pBluescript. To construct the targeting vector, a 6.7-kb SalI fragment of the plasmid pGT1.8IresBgeo containing the En2-derived splice acceptor and the IresLacZ-Neo fusion gene (19) was subcloned into the SalI site of pB529/64. The final construct is shown in Fig. 1A.
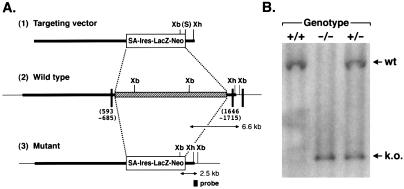
Figure 1.
Targeted disruption of the Cry1 gene. (A) Targeting of the Cry1 locus. The construct (1) was used to target the Cry1 gene (2) in the E14 g2a embryonic stem cell line. Homologous recombination leads to the deletion of a 13-kb genomic region [stippled box in (2)] containing exon sequences encoding the FAD-binding domain. The targeted allele (3) is detected by a probe as shown, with the expected DNA fragment sizes as indicated. Solid boxes, identified coding sequences; SA, Engrail-2 splice acceptor; S, SalI; Xb, XbaI; Xh, XhoI (restriction sites). (B) Identification of targeted mutants by Southern hybridization. The 2.5-kb mutant and the 6.6-kb wild-type fragments resulting from Xba digestion are indicated.
The targeting vector (100 μg) was linearized with XhoI and electroporated into E14tg2a embryonic stem cells (20). Electroporated cells were cultured in the presence of 150 μg/ml G418 (GIBCO) for 8 days; G418-resistant clones were picked and genotyped by Southern hybridization. One embryonic stem clone containing the disrupted Cry1 gene was injected into a C57BL/6 blastocyst and transferred into pseudopregnant female recipients. The resulting chimeras were bred with C57BL/6 females. Germ-line transmission was screened by coat color and confirmed by Southern hybridization. By crossing heterozygotes generated by gene targeting we obtained Cry1−/− mice.
Generation of Cry1−/−Cry2−/− Mice.
The Cry1−/− mice were crossed with Cry2−/− mice (12) to obtain Cry1+/−Cry2+/− progeny. The double heterozygotes then were interbred to obtain Cry1−/−Cry2−/− animals as well as double heterozygotes. Genotyping was done both by Southern hybridization and by PCR by using two sets of primers (one for amplifying the wild type and the other for amplifying the disrupted gene) for each of the Cry genes. After verifying the reliability of the PCR genotyping, all routine screening was done by PCR.
Locomotor Activity Analysis.
The experimental setup for recording locomotor activity (wheel running) has been described previously (21–23). Activity data was recorded continuously by a PC system (Chronobiology Kit; Stanford Software Systems, Santa Cruz, CA) and was displayed and analyzed by using software in the MatLab environment (ClockLab; Actimetrics, Evanston, IL). The period in DD was measured by a χ2 periodogram from 20 consecutive days when no manipulations were performed (24). Fast Fourier transform (FFT) analysis of the same 20 consecutive days also was performed (25). The power spectral densities for frequencies ranging from 0 to 1 cycle/hr were determined and normalized to a total power (area under the curve) of 1.0. This normalization, in part, corrects for individuals’ differences in activity level. The peak in the circadian range (18- to 30-hr period or 0.033–0.055 cycles/hr) of the relative power was determined for each animal for comparison. Effects of genotype were analyzed by a Generalized Model (GLM) ANOVA by using ncss (Kayesville, UT), with Scheffe’s posthoc tests for pairwise comparisons.
In Situ Hybridization.
The oscillation of mPer1 and mPer2 mRNA levels in the SCN of mutant mice under LD, DD, and acute light induction conditions was quantified by in situ hybridization as described previously (26). Animals were killed by cervical dislocation, and brains were dissected rapidly and frozen in dry ice. For analysis of diurnal expression and acute light induction, animals were sacrificed at ZT6 and ZT18, the midpoints of the light and dark phases, respectively. At ZT18, mice were either sacrificed in dim (15-W) yellow light without receiving a light pulse or were exposed to a 1-hr white fluorescent light pulse (30 W, positioned 4 inches above animals) beginning at ZT18 and returned to darkness for a half-hour after the end of the light pulse before sacrifice. For analysis of expression in DD, animals were transferred to constant darkness at the usual lights-off time and then sacrificed 22 or 34 hr later. Mice were dislocated cervically, and the optic nerves were cut by using an infrared viewer (FJW Industries, Palatine, IL); dissections were completed with dim, red (15-W, Kodak safelamp filter 1A) illumination. 33P[UTP]-labeled riboprobes were generated from template DNA containing nucleotides 340–761 (GenBank accession no. AF022992) and 9–489 (GenBank accession no. AF035830) for mPer1 and mPer2, respectively. Sections (20 μm) throughout the extent of the SCN were collected such that alternate sections from the same brains were hybridized for mPer1 and mPer2. Autoradiograms (Kodak BioMax MR) were digitized by using a Polaroid SprintScan slide scanner and optical densities were quantitated by using nih image software on a Macintosh computer. Signals were calibrated by comparison of optical densities obtained from the same film exposed to 14C radioactive standards (American Radiolabeled Chemicals, St. Louis) inserted into each film cassette. Effects of genotype, time, or conditions or their interactions were analyzed by ANOVA using NCSS, with Scheffe’s test for posthoc pairwise comparisons.
Results
Generation of Cry1−/− and Cry1−/−Cry2−/− Mice.
The Cry2−/− mice have been described previously (12). The Cry1−/− were generated by the same approach using the targeting strategy summarized in Fig. 1A. The transmission of the mutated gene was determined by Southern hybridization. The original heterozygotes obtained by gene targeting then were crossed with C57BL/6 to obtain Cry1+/− mice. The Cry1−/− mice were obtained by mating these heterozygotes. Genotyping was done by both Southern hybridization and PCR. Fig. 1B shows the results of a genotyping assay by Southern hybridization. The Cry1−/− mice were represented at the expected Mendelian frequency and, hence, appear to have no survival disadvantage.
To obtain Cry1−/−Cry2−/− mice, the Cry1 heterozygotes were crossed with Cry2−/− mice to obtain Cry1−/+Cry2−/+ progeny. The double heterozygotes were then interbred to obtain Cry1−/−Cry2−/− animals (Fig. 2). The double homozygous mutants, as well, occurred at the predicted frequency and appeared physically indistinguishable from their wild-type littermates.
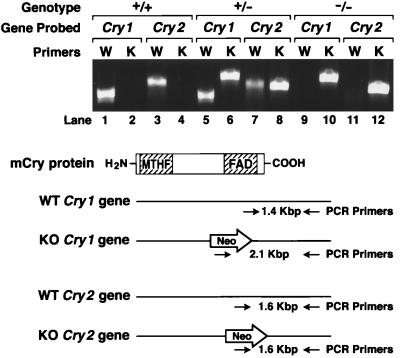
Figure 2.
Genotyping of progeny from Cry1+/−Cry2+/− cross by PCR. The knockouts of both genes each were generated by deleting a segment of the wild-type gene encoding the FAD-binding domain of CRY1 (amino acids 230–549) and of CRY2 (amino acids 349–569) and replacing it with the Neo gene. As shown in the schematic diagram, primers hybridizing to the deleted region were used to detect the wild type and primers hybridizing to the Neo gene were used to detect the mutated genes. The photograph shows results of PCR analysis of wild-type, double heterozygous, and double homozygous mutant mice.
Circadian Behavior of Cry1−/− and Cry1−/−Cry2−/− Mice.
In previous work, Cry2−/− mice were found to have abnormal circadian behavior, but were still capable of entrainment to light (12). It was suggested that the photic response in these mutants was mediated by either the CRY1 protein or a nonrod and noncone opsin (12). Hence, it was of interest to determine the effects of the Cry1−/− genotype both with regard to the free-running period and entrainment to light/dark cycles. Fig. 3 A and B shows that under LD12:12 conditions the mutants are indistinguishable from wild-type littermates, consistent with normal photoentrainment. Interestingly, as in the case of Cry2−/− mice, the Cry1−/− animals exhibit an altered free-running period under DD conditions (Fig. 3B). However, in contrast to Cry2−/− mice (Fig. 3C), which have a free-running period 1 hr longer than normal (Fig. 3A), Cry1−/− mice exhibit significantly shorter free-running periods than wild type (Fig. 3E; GLM ANOVA, F(5) = 12.2, P < 1 × 10−7; Scheffe’s posthoc comparison, P ≤ 0.05). Thus, it appears that both CRY1 and CRY2 proteins can influence the steady-state periodicity of the circadian system in mice, but apparently “pull” the oscillator in opposite directions.
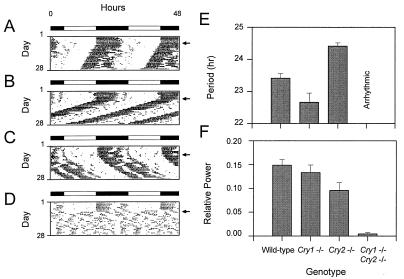
Figure 3.
Effects of disruption of the Cry genes on circadian locomotor activity rhythms. (A–D) Wheel-running activity records of individual mice, double-plotted according to convention so that each day’s data are represented both to the right and beneath that of the preceding day. Times of activity are represented by black. The animals were kept on a LD12:12 cycle as indicated by the bar above each record and then transferred to constant darkness by allowing lights to go off at the usual time on the day, indicated by an arrow on the right. (A) Activity record of a wild-type mouse. (B) Activity record of a Cry1−/− mouse. (C) Activity record of a Cry2−/− mouse. (D) Activity record of a Cry1−/−Cry2−/− mouse. (E) Effects of disruption of the Cry genes on circadian period. The free-running period was estimated by χ2 periodogram from days 1–20 in DD. Means and SEM of each genotype are illustrated. Sample sizes (N) are as follows: wild-type, n = 6; Cry1−/−, n = 5; Cry2−/−, n = 10; Cry1−/−Cry2−/−, n = 4. None of the double homozygotes exhibited significant circadian periodicity and, hence, no period estimates are shown. (F) Loss of circadian rhythmicity was assessed by Fourier analysis. Data from days 1–20 in DD were analyzed by fast Fourier transform (FFT), and power spectral densities for frequencies ranging from 0 to 1 cycles/hr were determined and normalized to a total power (area under the curve) of one. The resultant relative power value peak in the circadian range (18- to 30-hr period or 0.033- 0.055 cycles/hr) was determined for each animal for comparison. The means and SEM of circadian peak values of relative power are plotted for each genotype. Sample sizes are the same as in E.
Because CRY1 and CRY2 mutants are still capable of entrainment to light, we analyzed mice lacking both cryptochromes. The locomotor activity rhythms of Cry1−/−Cry2−/− mice (Fig. 3D) under LD12:12 conditions appeared normal. However, upon transfer to DD, the double mutants became arrhythmic immediately (Fig. 3 D and F). Some weakening in circadian rhythmicity is also apparent in Cry2−/− mice. For comparison, activity records of Cry2−/− mice kept under identical conditions were subject to the same FFT analysis as the individuals in the present study. A significant reduction in relative power in the circadian range is seen in Cry2−/− mice compared with wild-type mice (GLM ANOVA, F(6) = 2.81, P = 0.02; Scheffe’s posthoc comparison, P ≤ 0.05). Yet, a more substantial reduction in circadian power is observed in the double mutants (Fig. 3F; GLM ANOVA, F(6) = 2.81, P = 0.02; Scheffe’s posthoc comparison, P ≤ 0.05). These results show that, jointly, the two cryptochromes are essential for the expression of sustained circadian rhythms of locomotor behavior in vivo. The complete lack of residual circadian rhythmicity in DD is striking and differs from that seen in Clock and mPer2 mutant mice (21, 23, 27).
Status of the Molecular Clock in Cry1−/− and Cry1−/−Cry2−/− Mutants.
The central role of cryptochromes in controlling behavioral circadian rhythms makes it difficult to distinguish their putative role as photoreceptors from a role within the core oscillator itself. Hence, we wished to analyze the circadian pacemaker in the SCN and its response to light at the molecular level to address this issue. Currently, the known likely components of the molecular oscillator in mouse are Clock (21–23), BMAL1 (28–29), and mPer1, mPer2, mPer3, and mTim (26, 27, 30–32), of which mPer1 and mPer2 show robust circadian oscillations of gene expression and light induction in the SCN (see ref. 3). Hence, we examined the oscillations of mPer1 and mPer2 transcript levels and their induction by acute light pulses to assess the roles of CRY1 and CRY2 proteins in photoreception as well as in the intrinsic oscillations of the molecular clock.
Fig. 4 A and B shows the effects of Cry genotype on mPer1 expression in the SCN under LD12:12 and in response to an acute light pulse at ZT18. As would be expected, mPer1 levels were elevated significantly during the day (ANOVA, F(1) = 20.02, P < 0.005) and in response to light pulses (ANOVA, F(1) = 6.25, P < 0.05). Significant genotype by light-condition interactions were detected (ANOVA, F(2) = 5.56, P < 0.05), reflective of differential response to light among the genotypes. In particular, Cry1−/− mice had diminished light induction at ZT18 (only ≈30% of wild type; Scheffe’s posthoc comparison, P ≤ 0.05) and Cry1−/− Cry2−/− mutants had significantly elevated nocturnal levels with no significant induction in response to light (Scheffe’s posthoc comparison, P ≤ 0.05). The nocturnal elevation of mPer1 mRNA levels is consistent with a light-independent negative regulation by CRY1 and CRY2. In addition, the absence of an acute light response of mPer1 suggests either that CRY1 and CRY2 are required for this photoresponse or that there is a ceiling effect on mPer1 induction.
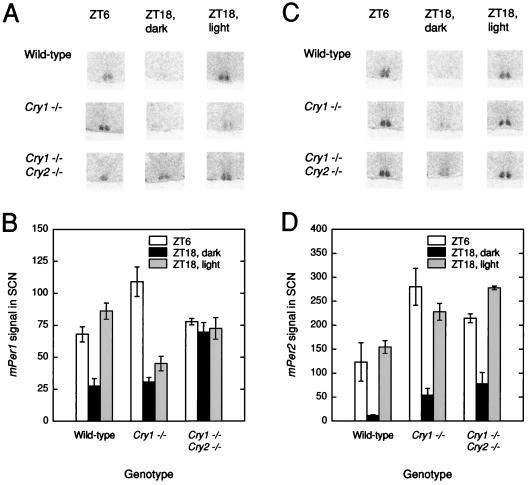
Figure 4.
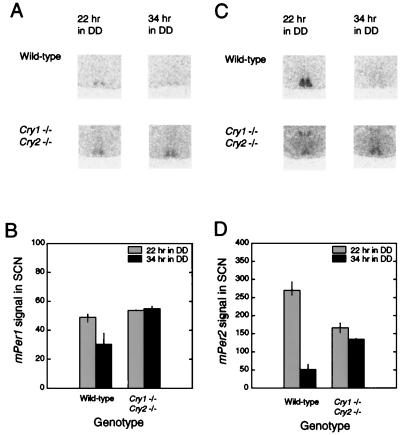
Effects of disruption of the Cry genes on diurnal expression of mPer1 and mPer2. (A) Representative in situ mPer1 signal in the SCN regions of mice of three different genotypes and under three different conditions. (B) Means and ranges in signal values for mPer1 in the SCN by time/light condition and genotype. (C) Representative in situ mPer2 signal in the SCN regions of mice of three different genotypes and under three different conditions. (D) Means and ranges in signal values for mPer2 in the SCN by time/light condition and genotype. n = 2 per genotype per condition.
The mPer2 results, shown in Fig. 4 C and D, were quite different. As with mPer1, significant elevation during the day (ANOVA, F(1) = 57.38, P = 0.0003) or in response to an acute light pulse (ANOVA, F(1) = 210.2, P = 0.000007) was present. However, the day–night differences and photic induction were exhibited by all three genotypes, so that no significant genotype interaction was detected by ANOVA. However, a significant main effect of genotype was present (ANOVA, F(2) = 21.52, P = 0.002) because of elevated levels in all conditions in the Cry1−/− Cry2−/− mutants relative to wild type (Scheffe’s posthoc comparison, P < 0.05). Thus, there is an unexpected difference in the regulation of mPer1 and mPer2 in Cry double mutants.
To determine whether the day–night difference in mPer2 was a light-driven or true circadian oscillation, we sought to determine whether either or both of the mPer transcripts were continuing to oscillate in DD in the Cry1−/− Cry2−/− mice. Two time points in DD (22 and 34 hr, or approximately CT 10 and 22) were selected corresponding to the peak and trough times for mPer2 in the SCN of wild-type mice. As shown in Fig. 5 A and B, in the Cry1−/− Cry2−/− mice, there is no apparent oscillation of mPer1 in DD and levels are constitutively elevated, consistent with the observations under LD conditions. ANOVA revealed significant genotype, time, and genotype × time interactions (GLM ANOVA; genotype: F(1) = 28.64, P < 0.002; time: F(1) = 10.13, P < 0.02; genotype × time: F(1) = 12.97, P < 0.02). However, the results with mPer2 expression in DD conditions contrasted to those seen in response to light and were similarly invariant as mPer1 but at intermediate values (Fig. 5 C and D). Because the mPer2 mean values of the mutants were not different from wild types, ANOVA failed to detect a significant genotype effect; however, significant time effects and time × genotype interactions were detected (GLM ANOVA; genotype: F(1) = 0.98, P = 0.36; time: F(1) = 149.2, P < 0.00002; genotype × time: F(1) = 83.09, P < 0.0001). Thus, the lack of circadian oscillations in mPer1 and mPer2 gene expression in Cry1−/− Cry2−/− is consistent with behavioral arrhythmicity in these animals under DD conditions (Fig. 3D).
Figure 5.
Effects of disruption of the Cry genes on circadian expression of mPer1 and mPer2. (A) Representative in situ mPer1 signal in the SCN regions of wild-type and Cry1−/−Cry2−/− mutant mice and at two times in DD. (B) Means and ranges in signal values for mPer1 in the SCN by time and genotype. (C) Representative in situ mPer2 signal in the SCN regions of wild-type and Cry1−/−Cry2−/− mutant mice and at two times in DD. (D) Means and ranges in signal values for mPer2 in the SCN by time and genotype. n = 3 wild types; two mutants per condition.
Discussion
Our results raise a number of issues regarding the potential role of cryptochromes as circadian photoreceptors and as integral components of the molecular clock. It is still difficult to place the cryptochromes definitively within the pathways that comprise the circadian system; however, with the information at hand, certain provisional conclusions can be made.
Cryptochromes as Circadian Photoreceptors.
Elimination of either CRY1 or CRY2 protein reduces and elimination of both abolishes the acute light inducibility of mPer1 in the SCN. Both sensitivity and phase of light induction of mPer1 correlate with behavioral phase shifts, leading to the proposal that this may be a mechanism by which light resetting of the circadian clock occurs (33). Considering the photoreceptor function of all other members of their class of proteins (5, 6), the simplest interpretation is that CRY1 and CRY2 are circadian photoreceptors. In agreement with this notion, genetic evidence in A. thaliana (14) and Drosophila (13, 15, 16) show that cryptochromes act as circadian photoreceptors in these organisms. Furthermore, it is now well established that mice lacking rod and cone photoreceptor cells have normal circadian photoentrainment (34–36), which is consistent with the claim that cryptochromes in the ganglion cells and inner nuclear layer of the retina are the circadian photoreceptors (8). However, the diurnal variation of mPer2 in the SCN and its normal inducibility by acute light pulses, combined with the finding that mPer2 may be a component of the circadian clock (27), would seem to indicate that there are at least two photic input pathways into the clock as has been proposed for Drosophila (13). Indeed, in recent years a number of nonrod, noncone opsins have been identified in mammals (see ref. 37) that conceivably could act as circadian photoreceptors in rodless and coneless mice. Clearly, there will be no formal proof that cryptochromes are circadian photoreceptors until the photoreception and phototransduction mechanisms are understood at molecular detail.
The apparent rhythmicity, both behavioral and in mPer2 expression in Cry1−/−Cry2−/− mice under LD conditions, raises the question of whether this can be more appropriately termed entrainment or “masking.” Masking refers to the obscuring of the period phase, or presence/absence of a circadian rhythm, by the response to a light pulse or other environmental Zeitgeber. In a nocturnal rodent, activity onset may be delayed until lights-off, for example, by the animal’s reluctance to move about in a brightly lit environment. In a narrow sense, the term masking would seem not to apply because there is no need to assume the behavior is driven by factors other than the clock mechanism; after all, mPer2 expression continues to change. However, entrainment also seems inappropriate because both the behavioral and mPer2 rhythms seem to be abolished within the first cycle in constant conditions. A more appropriate term, hence, seems to be “light-driven.” The correspondence between the mPer2 expression and the behavioral rhythm could indicate that mPer2 is more closely involved in driving behavior than other clock genes. If this were so, then a light-driven mPer2 rhythm could, in turn, drive a behavioral rhythm.
Cryptochromes as Clock Components.
The data presented here and by others (17) provide in vivo evidence that cryptochromes play a central role within the mammalian circadian clock system. First, mutations in either Cry1 or Cry2 alter the length of the circadian period in mice in vivo. Second, in Cry1−/−Cry2−/− double mutants there is no behavioral rhythmicity and no molecular oscillation of either mPer1 or mPer2, which are thought to be components of the molecular clock (31, 32). Finally, Cry1, like many of the other clock genes, is expressed with a circadian rhythm in the SCN (8). Whether the behavioral arrhythmicity seen at the organismal level is a cell-autonomous effect in SCN neurons (4) or is the result of desynchrony of SCN cellular oscillators remains to be determined. In either case, it is conceivable that the cryptochromes could exert their circadian phenotypic effects at the level of input pathways to the circadian oscillator system; however, the molecular effects on mPer gene expression suggest a more central role. One caveat here is that in Drosophila, which has a circadian photoreception system similar to those of mammals (1, 3), it has been reported that the absence of cryptochrome affects photoentrainment but not the free-running period (13). However, the mutant used in that study had a conservative change (Asp → Asn) at the flavin-binding site (13). Conceivably, this mutation could abolish the photoreceptor function but not affect the light-independent clock function.
Model for the Mammalian Clock.
We have made three striking molecular observations in this study: (i) the levels of mPer1 and mPer2 transcripts in Cry1−/−Cry2−/− are elevated under LD conditions; (ii) the absence of diurnal variation of mPer1 but near-normal variation of mPer2 in the double mutant under LD; and (iii) the absence of apparent oscillations of both mPer1 and mPer2 in CRY double mutants under DD conditions. These results are consistent with cryptochromes playing a photoreceptive role as well as acting as regulators of mPer gene expression in light-dependent (Fig. 4) and light-independent (Fig. 5) pathways.
That the basal levels of mPer1 increase in Cry1−/− and Cry1−/−Cry2−/− mice and that overall levels of mPer2 in LD are elevated in Cry1−/−Cry2−/− suggest that CRY proteins inhibit mPer gene expression. This could occur by one of the following mechanisms: CRY1 and 2 proteins could be repressors of the mPer genes; CRY1 and 2 proteins could interact with CLOCK or BMAL1 and interfere with the positive activation of mPer transcription; or CRY1 and CRY2 could interact with PER and TIM proteins and interfere with their negative feedback on the CLOCK-BMAL1 complex. At present, we cannot differentiate among these possibilities. However, by using recombinant CRY1 and CRY2 proteins, we failed to detect interaction with the E box regulatory element of the mPer1 gene and with the CLOCK protein made by in vitro transcription/translation. However, we did detect weak interactions of CRY with TIM and PER proteins made by in vitro transcription/translation (C.P.S., E. Vagas, and A.S., unpublished data). In a recent cotransfection study, Kume et al. (38) present data indicating that PER-CRY can negatively affect CLOCK-BMAL1-driven mPer1 transcription. Our results provide in vivo evidence of such a negative feedback role of Cry genes on mPer1. The damping of mPer2 expression in Cry1−/−Cry2−/− mutants toward an intermediate value could indicate either a diminished negative feedback on mPer2 as compared with mPer1 or that the Cry genes’ influence on mPer2 is offset by some positive influence. The differential effect of the two Cry genes on period raises the possibility that they have differential effects on mPer2 as well.
In either case, it is apparent that the Cry genes differentially influence mPer1 and mPer2 transcription. Differences in both the timing and the phases at which light can induce these transcripts in the SCN (30, 39) and, apparently, the phenotype of the null (27) all suggest that these two genes have nonredundant functions. To what extent their regulation by cryptochromes are involved in the differences in the functions of mPer1 and mPer2 remains to be determined.
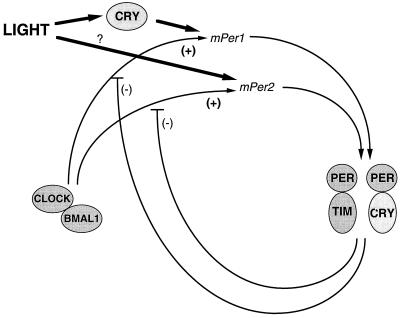
Thus, taken together, we propose the working model for the mammalian clock system shown in Fig. 6. CRY1 and CRY2 could participate in the circadian transcription–translation feedback loop and, in addition, act as the dominant photoreceptive elements for mPer1 acute light responses. In addition, there is photic input by another photoreceptor, which either directly or indirectly induces mPer2 acute light responses. Within the circadian feedback loop, both CRYs could interact with the other clock proteins, such as PER and TIM, to provide negative feedback (because in their absence mPer levels are elevated). The two CRY proteins may work synergistically in some roles (e.g., in their role related to the expression of circadian behavioral and mPer rhythms) and antagonistically in others (e.g., affecting the free-running period in opposite directions). Clearly more work is needed to develop a more specific model. This awaits the elucidation of the significance of the interaction of CRY1 and 2 with other signaling molecules such as phosphoprotein phosphatase 5 (40) as well as development of an in vitro assay for the photochemical reaction catalyzed by cryptochrome.
Figure 6.
The dual role of cryptochromes in the circadian clock. A model of genetic interactions among cryptochromes and other elements in the circadian autoregulatory loop are shown. A presumed basic feedback loop of positive mPer transcriptional drive of CLOCK-BMAL1 inhibited by PER-TIM is depicted. Cryptochromes appear to mediate light induction of mPer1 but not mPer2 (left side). In addition, the potential for cryptochromes to dimerize with PERs suggests that they may both function in a negative feedback as well (right side).
Acknowledgments
We thank Dr. David P. King for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants GM31082 (A.S.) and AG11412 (J.S.T.), the National Science Foundation Center for Biological Timing (J.S.T.), and a grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan (T.T.). J.S.T. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- Cry1 and Cry2 and CRY1 and CRY2
cryptochrome 1 and 2 genes and proteins, respectively
- mPer1 and mPer2
mouse period genes 1 and 2, respectively
- LD12:12
12-hr light/12-hr dark
- DD
constant darkness
- ZT
Zeitgeber time
- CT
circadian time
- SCN
suprachiasmatic nucleus
References
- 1.Young M. Annu Rev Biochem. 1998;37:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 2.Green C B. Trends Cell Biol. 1998;8:224–230. doi: 10.1016/s0962-8924(98)01269-0. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap J C. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Herzog E D, Takahashi J S, Block G D. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 5.Cashmore A R. J Plant Res. 1998;111:267–270. [Google Scholar]
- 6.Todo T. Mutat Res. 1999;434:89–97. doi: 10.1016/s0921-8777(99)00013-0. [DOI] [PubMed] [Google Scholar]
- 7.Hsu D S, Zhao X, Zhao S, Kazantsev A, Wang R P, Todo T, Wei Y F, Sancar A. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 8.Miyamoto Y, Sancar A. Proc Natl Acad Sci USA. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todo T, Ryo H, Yamamoto K, Hoh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 10.Van der Spek P J, Kobayashi K, Bootsma D, Takao M, Eker A P M, Yasui A. Genomics. 1996;27:177–182. doi: 10.1006/geno.1996.0539. [DOI] [PubMed] [Google Scholar]
- 11.Todo T, Tsuji H, Oteshi E, Hitomi K, Kim S T, Ikenaga M. Mutat Res. 1997;384:195–120. doi: 10.1016/s0921-8777(97)00032-3. [DOI] [PubMed] [Google Scholar]
- 12.Thresher R J, Vitaterna M H, Miyamoto Y, Kazantsev A, Hsu D S, Petit C, Selby C P, Dawut L, Smithies O, Takahashi J S, et al. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 13.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay S A, Rosbash M, Hall J C. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 14.Somers D, Devlin P, Kay S A. Science. 1998;282:488–490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 15.Emery P, So W V, Kaneko M, Hall J C, Rosbash M. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa T, Matsumoto A, Kato T, Jr, Togashi S, Ryo H, Ikenaga M, Todo T, Ueda R, Tanimura T. Genes Cells. 1999;4:57–66. doi: 10.1046/j.1365-2443.1999.00237.x. [DOI] [PubMed] [Google Scholar]
- 17.Van der Horst G T J, Muijtjens M, Kobayashi K, Tokano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker A P M, van Leenen D, et al. Nature (London) 1999;348:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 18.Park H W, Kim S T, Sancar A, Deisenhofer J. Science. 1995;268:1868–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 19.Mountford P, Zebnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooper M L, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 21.Vitaterna M H, King D P, Chang A M, Kornhauser J M, Lowrey P L, McDonald J D, Dove W F, Pinto L H, Turek F W, Takahashi J S. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M O, Steeves T D, Vitaterna M H, Kornhauser J M, Lowrey P L, Turek F W, Takahashi J S. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antoch M P, Song E-J, Chang A-M, Vitaterna M H, Zhao Y, Wilsbacher L D, Sangoram A M, King D P, Pinto L H, Takahashi J S. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokalove G, Bushell W. J Theor Biol. 1978;72:131–160. doi: 10.1016/0022-5193(78)90022-x. [DOI] [PubMed] [Google Scholar]
- 25.Bracewell R N. The Hartley Transform. New York: Oxford Univ. Press; 1986. [Google Scholar]
- 26.Sangoram A M, Saez L, Antoch M P, Gekakis N, Stakins D, Whiteley A, Fruechte E M, Vitaterna M H, Shimomura K, King D P, et al. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 27.Zheng B, Larkin D M, Albrecht V, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. Nature (London) 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 28.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz J C. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 29.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albrecht U, Sun Z, Eichele G, Lee C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 31.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 32.Zylka M, Shearman L, Weaver D, Reppert S. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 33.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros J J, Dunlap J C, Okamura H. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 34.Foster R G, Provencio I, Hunson D, Fiske S, de Grip W, Menaker M. J Comp Physiol. 1991;169:39–50. doi: 10.1007/BF00198171. [DOI] [PubMed] [Google Scholar]
- 35.Provencio I, Wong S, Lederman A, Argamaso S M, Foster R G. Vision Res. 1994;34:1799–1806. doi: 10.1016/0042-6989(94)90304-2. [DOI] [PubMed] [Google Scholar]
- 36.Freedman M S, Lucas R J, Foster R G. Science. 1999;284:502–504. doi: 10.1126/science.284.5413.502. [DOI] [PubMed] [Google Scholar]
- 37.Blackshaw S, Synder S H. J Neurosci. 1999;19:3681–3690. doi: 10.1523/JNEUROSCI.19-10-03681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X, Maywood E S, Hastings M H, Reppert S M. Cell. 1999;98:193–206. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 39.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 40.Zhao S, Sancar A. Photochem Photobiol. 1997;66:727–731. doi: 10.1111/j.1751-1097.1997.tb03214.x. [DOI] [PubMed] [Google Scholar]