Abstract
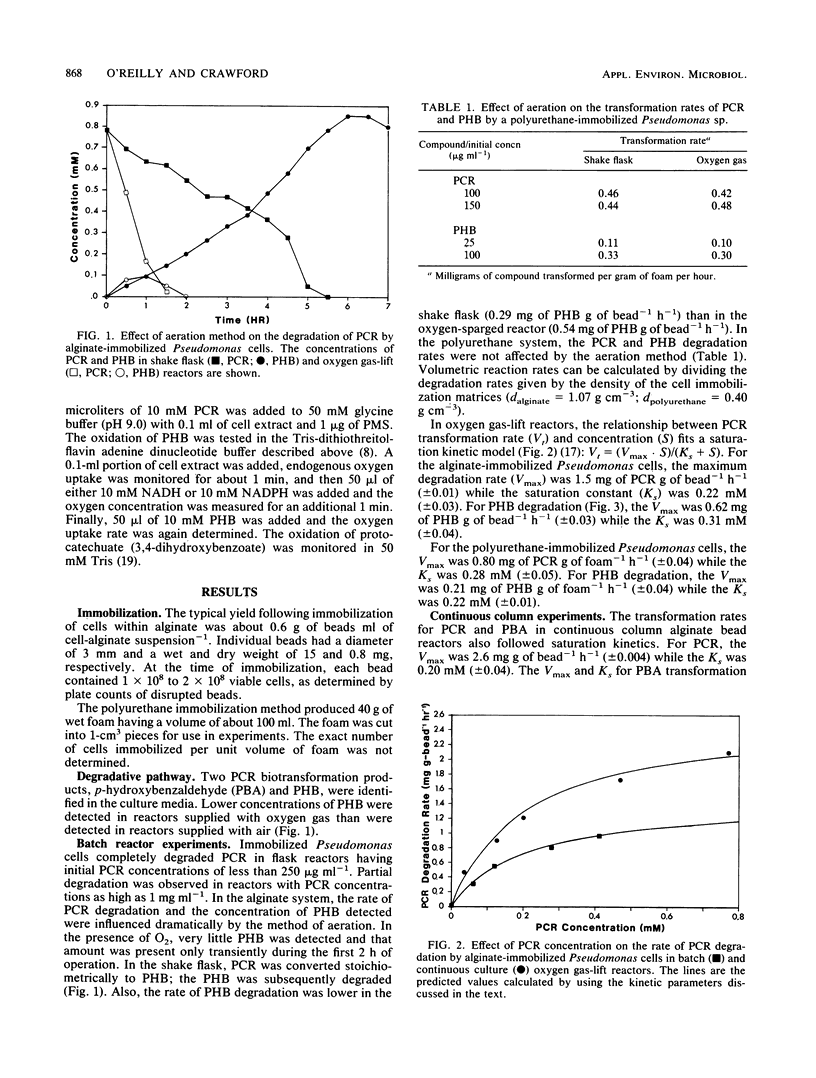
A p-cresol (PCR)-degrading Pseudomonas sp. was isolated from creosote-contaminated soil and shown to degrade PCR by conversion to protocatechuate via p-hydroxybenzaldehyde (PBA) and p-hydroxybenzoate (PHB). Cells of the Pseudomonas sp. were immobilized in calcium alginate beads and in polyurethane foam. The relationship between the PCR concentration and the PCR transformation rate was investigated in batch and continuous culture bioreactors. The biodegradation kinetics of PBA and PHB also were investigated. In batch culture reactors, the maximum PCR degradation rate (Vmax) for the alginate-immobilized Pseudomonas sp. cells was 1.5 mg of PCR g of bead-1 h-1 while the saturation constant (Ks) was 0.22 mM. For PHB degradation, the Vmax was 0.62 mg of PHB g of bead-1 h-1 while the Ks was 0.31 mM. For polyurethane-immobilized Pseudomonas sp. cells, the Vmax of PCR degradation was 0.80 mg of PCR g of foam-1 h-1 while the Ks was 0.28 mM. For PHB degradation, the Vmax was 0.21 mg of PHB g of foam-1 h-1 and the Ks was 0.22 mM. In a continuous column alginate bead reactor, the Vmax for PCR transformation was 2.6 mg g of bead-1 h-1 while the Ks was 0.20 mM. The Vmax and Ks for PBA transformation in the presence of PCR were 0.93 mg g of bead-1 h-1 and 0.063 mM, respectively. When PHB alone was added to a reactor, the Vmax was 1.48 mg g of bead-1 h-1 and the Ks was 0.32 mM.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DAGLEY S., PATEL M. D. Oxidation of p-cresol and related compounds by a Pseudomonas. Biochem J. 1957 Jun;66(2):227–233. doi: 10.1042/bj0660227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer D. F., Krumme M. L., Boyd S. A., Tiedje J. M. Kinetics of phenol biodegradation by an immobilized methanogenic consortium. Appl Environ Microbiol. 1986 Aug;52(2):345–351. doi: 10.1128/aem.52.2.345-351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal A., Neujahr H. Y. Metabolism of phenol and resorcinol in Trichosporon cutaneum. J Bacteriol. 1979 Jan;137(1):13–21. doi: 10.1128/jb.137.1.13-21.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J. Incorporation of [18O]water in the formation of p-hydroxybenzyl alcohol by the p-cresol methylhydroxylase from Pseudomonas putida. Biochem J. 1978 Oct 1;175(1):345–347. doi: 10.1042/bj1750345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Taylor D. G. The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem J. 1977 Oct 1;167(1):155–162. doi: 10.1042/bj1670155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J. The hydroxylation of P-cresol and its conversion to P-hydroxybenzaldehyde in Pseudomonas putida. Biochem Biophys Res Commun. 1976 Mar 22;69(2):462–468. doi: 10.1016/0006-291x(76)90544-1. [DOI] [PubMed] [Google Scholar]
- Hosokawa K., Stanier R. Y. Crystallization and properties of p-hydroxybenzoate hydroxylase from Pseudomonas putida. J Biol Chem. 1966 May 25;241(10):2453–2460. [PubMed] [Google Scholar]
- Robinson J., Cooper J. M. Method of determining oxygen concentrations in biological media, suitable for calibration of the oxygen electrode. Anal Biochem. 1970 Feb;33(2):390–399. doi: 10.1016/0003-2697(70)90310-6. [DOI] [PubMed] [Google Scholar]
- Whittaker J. W., Lipscomb J. D., Kent T. A., Münck E. Brevibacterium fuscum protocatechuate 3,4-dioxygenase. Purification, crystallization, and characterization. J Biol Chem. 1984 Apr 10;259(7):4466–4475. [PubMed] [Google Scholar]


