Abstract
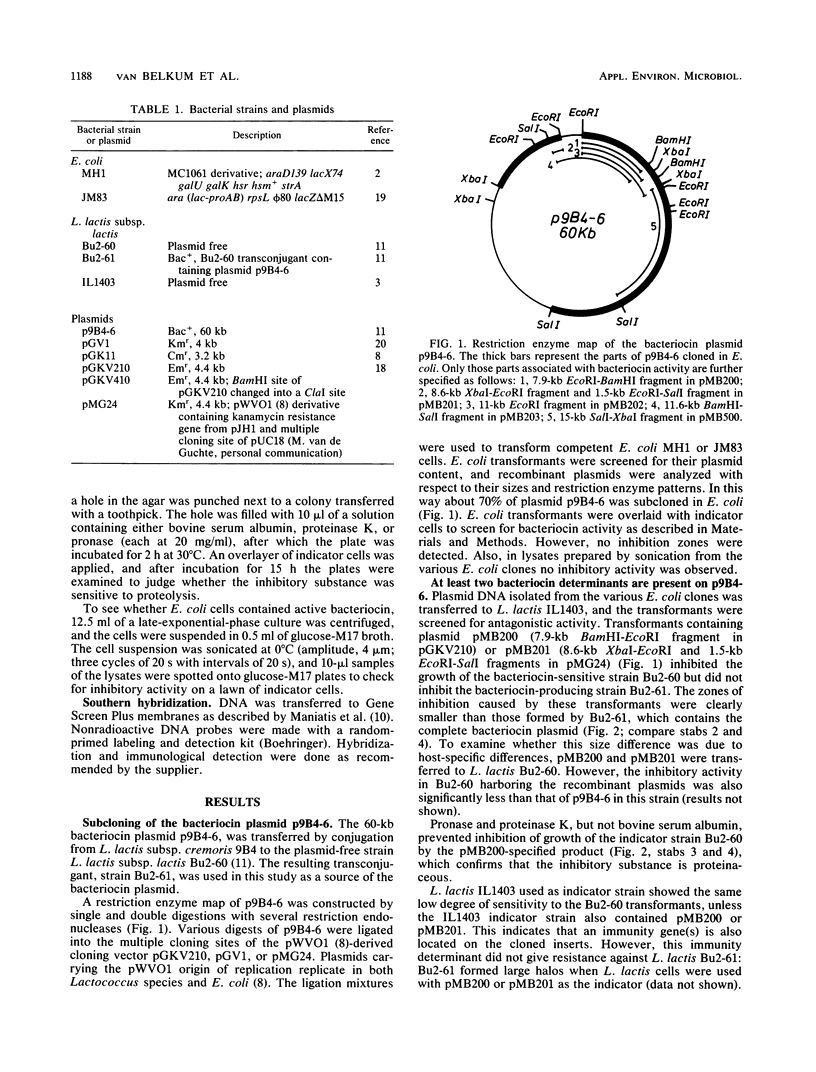
Lactococcus lactis subsp. cremoris 9B4 plasmid p9B4-6 (60 kilobases [kb]), which specifies bacteriocin production and immunity, was analyzed with restriction endonucleases, and fragments of this plasmid were cloned into shuttle vectors based on the broad-host-range plasmid pWVO1. Two regions on p9B4-6 were identified which specify inhibitory activity on L. lactis indicator strains: one that could be confined to a 1.8-kb ScaI-ClaI fragment with low antagonistic activity and a 15-kb XbaI-SalI fragment specifying high antagonistic activity. The inhibitory substances produced by these two clones were sensitive to proteolysis. A 4-kb HindIII fragment derived from the 15-kb fragment strongly hybridized with the 1.8-kb fragment. The antagonistic activity specified by the 4-kb fragment was somewhat reduced as compared with that of the 15-kb fragment. A 1.3-kb ScaI-HindIII subfragment of the 4-kb fragment contained both the immunity and bacteriocin genes. Inhibition studies showed that the two bacteriocins had different specificities.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Daeschel M. A., Klaenhammer T. R. Association of a 13.6-Megadalton Plasmid in Pediococcus pentosaceus with Bacteriocin Activity. Appl Environ Microbiol. 1985 Dec;50(6):1538–1541. doi: 10.1128/aem.50.6.1538-1541.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey G. P. Plasmid associated with diplococcin production in Streptococcus. Appl Environ Microbiol. 1984 Oct;48(4):895–896. doi: 10.1128/aem.48.4.895-896.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis A., Singh J., Teuber M. Potential of lactic streptococci to produce bacteriocin. Appl Environ Microbiol. 1983 Jan;45(1):205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon K. S., McKay L. L. Restriction enzyme analysis of lactose and bacteriocin plasmids from Streptococcus lactis subsp. diacetylactis WM4 and cloning of BclI fragments coding for bacteriocin production. Appl Environ Microbiol. 1987 May;53(5):1171–1174. doi: 10.1128/aem.53.5.1171-1174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R. Bacteriocins of lactic acid bacteria. Biochimie. 1988 Mar;70(3):337–349. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Neve H., Geis A., Teuber M. Conjugal transfer and characterization of bacteriocin plasmids in group N (lactic acid) streptococci. J Bacteriol. 1984 Mar;157(3):833–838. doi: 10.1128/jb.157.3.833-838.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Scherwitz K. M., Baldwin K. A., McKay L. L. Plasmid linkage of a bacteriocin-like substance in Streptococcus lactis subsp. diacetylactis strain WM4: transferability to Streptococcus lactis. Appl Environ Microbiol. 1983 May;45(5):1506–1512. doi: 10.1128/aem.45.5.1506-1512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vosman B., Kooistra J., Olijve J., Venema G. Cloning in Escherichia coli of the gene specifying the DNA-entry nuclease of Bacillus subtilis. Gene. 1987;52(2-3):175–183. doi: 10.1016/0378-1119(87)90044-8. [DOI] [PubMed] [Google Scholar]
- van der Lelie D., van der Vossen J. M., Venema G. Effect of Plasmid Incompatibility on DNA Transfer to Streptococcus cremoris. Appl Environ Microbiol. 1988 Apr;54(4):865–871. doi: 10.1128/aem.54.4.865-871.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]






