Abstract
Erythropoietin (EPO) controls the proliferation and differentiation of erythroid progenitor cells into red blood cells. EPO induces these effects by dimerization of the EPO receptors (EPOR) present on these cells. To discover nonpeptide molecules capable of mimicking the effects of EPO, we identified a small molecule capable of binding to one chain of EPOR and used it to synthesize molecules capable of inducing dimerization of the EPOR. We first identified compound 1 (N-3-[2-(4-biphenyl)-6-chloro-5-methyl]indolyl-acetyl-l-lysine methyl ester) by screening the in-house chemical collection for inhibitors of EPO binding to human EPOR and then prepared compound 5, which contains eight copies of compound 1 held together by a central core. Although both compounds inhibited EPO binding of EPOR, only compound 5 induced dimerization of soluble EPOR. Binding of EPO to its receptor in cells results in activation of many intracellular signaling molecules, including transcription factors like signal transducer and activator of transcription (STAT) proteins, leading to growth and differentiation of these cells. Consistent with its ability to induce dimerization of EPOR in solution, compound 5 exhibited much of the same biological activities as EPO, such as (i) the activation of a STAT-dependent luciferase reporter gene in BAF3 cells expressing human EPOR, (ii) supporting the proliferation of several tumor cell lines expressing the human or mouse EPOR, and (iii) the in vitro differentiation of human progenitor cells into colonies of erythrocytic lineage. These data demonstrate that a nonpeptide molecule is capable of inducing EPOR dimerization and mimicking the biological activities of EPO.
Erythropoietin (EPO) is essential for the maintenance of red blood cells in humans as well as in various animal models (1), reviewed in ref. 2. In humans, the kidney is the primary source of EPO synthesis, whereas other organs such as the liver and brain produce small but significant amounts in adults (3–6). Deficits in EPO production result in anemia in humans and in animal models. In humans, the most prevalent form of anemia is associated with kidney failure (7). At present, the only treatment for this form of anemia is administration of recombinant EPO via subcutaneous or intravenous injection (8–10). The use of recombinant EPO has significantly improved the quality of life of these patients; however, this treatment requires repeated administration of recombinant protein, which is both inconvenient and expensive.
EPO induces its biological effects after binding to a cell-surface receptor (EPOR). Binding of EPO to EPOR results in dimerization of these receptors, as is the case for many other growth factor and cytokine receptors (11, 12). Apparently dimerization of EPOR is all that is required to trigger the biological responses associated with EPO. A constitutively active (hormone-independent) EPOR was first isolated after retroviral transduction (13). The activation of this receptor mapped to an arginine-to-cysteine mutation at position 129 in the human EPOR. The mutant receptor forms disulfide-linked homodimers in the absence of EPO (14). After this example, more constitutively active EPORs have been created by introducing a cysteine residue in parts of the putative EPOR dimerization interphase (15, 16). These mutant receptors, when introduced into growth factor-dependent BAF3 cells, converted them into growth factor-independent cells. Similarly, a bivalent monoclonal antibody directed toward the extracellular domain of the EPOR promotes dimerization of EPOR and mimics EPO activities (17). Moreover, recently a 20-aa peptide, EPO mimetic peptide-1 (EMP-1), has been shown to dimerize the EPOR in solution as well as on the cell surface (18, 19). This peptide exhibits EPO-like activities both in vitro and in vivo (18).
The crystal structures of EPO/EPOR and EMP-1/EPOR complexes have been solved and reveal a different configuration of the EPOR dimer in each of the complexes (19, 20). On the basis of the three-dimensional structure of EPOR observed in these crystals, the mutations described above are in a region of the exoplasmic domain that is too far away for disulfide bond formation to occur between the two EPOR molecules. Therefore, it is unlikely that the covalently held EPOR dimers induced by mutations in the exoplasmic domain (as described above) will have a configuration similar to that of EPO- or EMP-1-induced EPOR dimers. A similar conclusion may be drawn for the monoclonal antibody-induced EPOR dimers. All these data suggest that, although the dimerization of the EPOR is important, the conformation of EPOR in the dimer complex is quite flexible. This also suggests that other molecules capable of dimerizing the EPOR may be able to act as EPO mimetics as well.
We are interested in developing small-molecule EPO mimetics for the treatment of anemia. On the basis of the characteristics of EPOR-dimerizing entities described above, it appears that such a molecule must have functional groups capable of interacting with at least two receptor chains. One possible way to obtain such a molecule is to first identify a compound that can interact with one chain of the EPOR and then ligate it in such a way that it can now interact with both chains of the receptor. In this paper, we report on the identification of such a molecule, which we initially identified as an EPOR antagonist. This molecule, when presented as an oligomer, is converted into an EPOR agonist, recapitulating some of the biological activities associated with EPO.
Materials and Methods
Synthesis of Compounds and EMP-1.
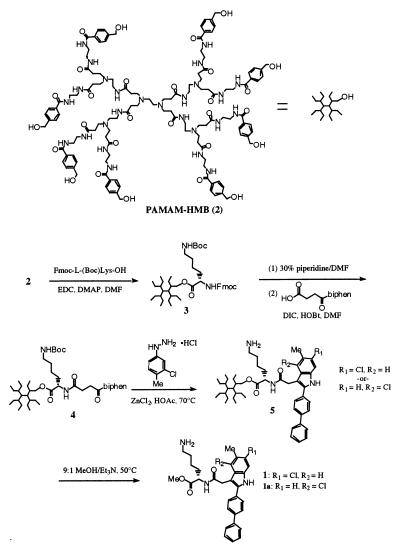
Compound 1 was synthesized by using Starburst polyamidoamino-octa-4-hydroxymethylbenzamide (2) as a soluble support, as shown in Fig. 1. To a stirring solution of support 2 (0.05 mmol, 105 mg) (21) and Fmoc-Lys(Boc)-OH (0.8 mmol, 364 mg, 2 eq per handle) in 3 ml of N,N-dimethylformamide (DMF) was added catalytic 4-dimethylaminopyridine (3–5 mg), followed by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (0.8 mmol, 154 mg). The reaction was stirred overnight at ambient temperature. Dendrimer 3 was isolated by size exclusion chromatography (SEC) on Sephadex LH-20 (Amersham Pharmacia) eluting with DMF, followed by SEC on Bio-Rad Biobeads S-X3 eluting with CH2Cl2. Removal of solvent in vacuo afforded 204 mg of 3 as a beige solid. Compound 3 (0.016 mmol, 100 mg) was treated with 3 ml of 30% piperidine in DMF for 15 min and isolated by SEC on Sephadex LH-20. The resulting octa-amine was dried in vacuo, and residue was dissolved in 3 ml of DMF containing 3-(4-biphenylbenzoyl)propionic acid (0.3 mmol, 76 mg, 2.3 eq) and 1-hydroxybenzotrazole hydrate (0.3 mmol, 41 mg). To the solution was added diisopropylcarbodiimide (0.3 mmol, 54 μL). The mixture was allowed to stand overnight, and the product was isolated by SEC on Biobeads S-X3 eluting with CH2Cl2. Removal of solvent afforded 114 mg of dendrimer 4 as a beige solid. Dendrimer 4 (84 mg) and 3-chloro-4-tolylhydrazinium chloride (1.5 mmol, 290 mg) were dissolved in 3 ml of glacial acetic acid containing zinc chloride (1.5 mmol, 204 mg) and anisole (0.6 mmol, 60 mg, 60 μL), and the slurry was heated overnight at 70°C. The solvent was removed in vacuo, and the residue was dissolved in DMF and purified by SEC on Sephadex LH-20. Removal of solvent afforded 68 mg of 5 as a tan solid. 1H NMR of 5 revealed the removal of the Boc-protecting group during the reaction. Dendrimer 5 (40 mg) was taken up in 2 ml of 9:1 MeOH/Et3N and heated at 50°C for 22 hr. The mixture was rotary evaporated to a tan film, which was taken up in MeCN. The insoluble dendrimer was filtered, and the light-colored filtrate was rotary evaporated to afford 17 mg of methyl esters 1 and 1b as the major products, in addition to the corresponding cyclized lactams. Separation of the two regioisomers by reverse-phase chromatography (C-8, H2O/MeCN gradient containing 0.15% trifluoroacetic acid) afforded 1.7 mg of N-3-[2-(4-biphenyl)-6-chloro-5-methyl]indolyl-acetyl-l-lysine methyl ester (1) and 2.7 mg of N-3-[2-(4-biphenyl)-4-chloro-5-methyl]indolyl-acetyl-l-lysine methyl ester (1a) as the trifluoroacetate salts. 1 1H NMR (400 MHz, CD3OD): 7.76 (d, 2H), 7.70 (d, 2H), 7.68 (d, 2H), 7.47 (t, 2H), 7.38 (t, 1H),7.27 (d, 1H), 7.06 (d, 1H), 4.58 (m, 1H), 4.04 (d, 2H), 3.72 (s, 3H), 2.87 (t, 2H), 2.43 (s, 3H), 1.85–1.98 (m, 1H), 1.57–1.70 (overlapping m, 3H), 1.43 (m, 2H). 1a 1H NMR (400 MHz, CD3OD): d7.78 (d, 2H), 7.75 (d, 2H), 7.68 (d, 2H), 7.48 (s, 1H), 7.46 (t, 2H), 7.41 (s, 1H), 7.36 (t, 1H), 4.52 (m, 1H), 3.86 (s, 2H), 3.70 (s, 3H), 2.81 (m, 2H), 2.45 (s, 3H), 1.90 (m, 1H), 1.73 (m, 1H), 1.63 (m, 2H), 1.40 (m, 2H). Dried powders were stored at ambient temperatures and used for preparation of stock solutions in 100% DMSO. Stock solutions prepared in this way were stored at −20°C until use and were used within a few months, during which no loss of activity was observed as determined by activities of these compounds in the EPOR-binding and luciferase-reporter gene assays described here. EMP-1, GGTYcyclo(CHFGPLTWVC)KPQGG-amide, was prepared by the solid-phase method (22) on a 431A Applied Biosystems peptide synthesizer and dissolved in 100% DMSO.
Figure 1.
Scheme 1 for the synthesis of compounds 1 and 5.
Cell Lines.
Murine BAF3 cells (kindly provided by Alan D’Andrea, Harvard Medical School, Boston), DA3 cells (kindly provided by Jim Ihle, St. Jude Children’s Research Hospital, Memphis, TN), and cell lines derived from these were maintained in RPMI-1640 supplemented with 10% FBS, antibiotic, l-glutamine (all from GIBCO/BRL), and 5 ng/ml mIL-3 (R & D systems) at 37°C in a humidified incubator. Human erythroleukemia cell line F36 (obtained from Riken Cell Bank, Tsukuba Science City, Japan) and TF-1 (obtained from K. Kitamura, Tokyo University, Tokyo) (23, 24) were maintained in the above-mentioned media supplemented with hIL-3 (5 ng/ml).
Plasmids.
pET15b/hEPOR-ECD expresses the extracellular domain of the human EPOR in Escherichia coli, and RcCMV/EPOR expresses human EPOR in mammalian cells and will be described elsewhere (H.M., unpublished data). pAH4-LUC contains six copies of the signal transducer and activator of transcription (STAT)-binding site from the interferon regulatory factor 1 gene cloned upstream of the herpes simplex virus–thymidine kinase minimal promoter and the luciferase gene (25). A gene cassette conferring resistance to the antibiotic Zeocine (Invitrogen) was introduced at a SalI site in the pAH4LUC to yield pAH4LUCZeo.
Expression and Purification of EPO-Binding Protein in E. coli.
E. coli strain BL21 DE3 (Novagen) containing pET15b/hEPOR-ECD was used for expression of the extracellular domain of the human EPOR. The resulting recombinant protein was processed to yield the recombinant EPO-binding protein (rEBP) as described previously (26). An EPO-dependent luciferase assay was used to further characterize the rEBP. The purified rEBP inhibited EPO-induced luciferase activity in this assay with an IC50 of 5–10 nM (S.Q. and R.R., unpublished data).
EPOR-Binding Assay.
One microgram of the purified His-tagged-rEBP in 70 μl of 1× PBS [1× PBS contains 137 mM NaCl, 2.68 mM KCl, 1.46 mM KH2PO4, 15.8 mM sodium phosphate, pH 7.2 (GIBCO/BRL)] containing 0.05% sodium azide was dispensed into 96-well high-binding microtiter plates (Costar 3922) and allowed to bind at 4°C for at least 24 hr. All unbound rEBP was removed by washing with 1× PBS containing 0.05% Tween 20 (Sigma), and nonspecific binding sites were blocked by incubating with 1% BSA (Pierce) in 1× PBS and 0.05% Tween 20. The plate-immobilized rEBP bound ([3-[125I]iodotyrosyl]) EPO (125I-EPO) with a Kd of ≈5 nM. All competition binding experiments were performed in a volume of 50 μl containing 5 nM 125I-EPO (Amersham, specific activity 300–900 Ci/mmol) in 1× PBS/0.5% BSA/0.05% Tween 20/0.01% sodium azide/5% DMSO +/− compounds. To set up these assays, each compound was diluted into the assay buffer (from the stocks that have been prepared in 100% DMSO) to yield the appropriate compound concentration and added to each well of a 96-well plate. These reactions were incubated at 4°C for 16–20 hr to allow equilibrium binding, and unbound 125I-EPO was removed by five rapid washes with cold 1× PBS containing 0.5% BSA. Bound radioactivity was counted, after addition of 100 μl of Microscint (Packard)/well, in a Topcount scintillation counter (Packard). Under these conditions, unlabeled EPO inhibited binding of 125I-EPO with an IC50 of ≈5 nM.
EPOR Dimerization Assay.
A detailed description of this assay will be published elsewhere (D.B. and S.Q., unpublished work). Briefly, rEBP containing a protein kinase A substrate site was produced in E. coli as described above. This protein was radiolabeled by using protein kinase A and 33P-γATP to high specific activity. The soluble 33P- rEBP was allowed to interact with the plate-bound rEBP as described for the receptor-binding assay. Each reaction was performed in a 100 μl volume containing 1- to 2-nM-labeled rEBP in 50 mM Hepes (pH 7.2)/5 mM Mg2Cl/5 mM Ca2Cl/0.05% sodium azide/1% BSA/5% DMSO/+/−compounds) in 96-well microtiter plates in the presence or absence of 250 ng/well immobilized rEBP. After incubation for 16–20 hr in the presence or absence of compounds, plates were washed, and the amount of radioactivity was determined as described in the EPOR-binding assay.
Establishment of Luciferase Reporter Cell Lines in BAF3 Cells.
The pAH4LUCZeo DNA was linearized at a PvuI site and transfected together with pHOOK3 (Invitrogen) into the BAF3 cells by electroporation. Forty-eight hours after transfection, the cells transfected with pHOOK3 were separated from the untransfected cells by using magnetic beads coated with phOx per manufacturer’s suggested protocol (Invitrogen). The isolated cells were plated at one cell per well in 96-well microtiter plates and allowed to grow in the presence of Zeocine (200 μg/ml) for selection of stable transfectants. Zeocine-resistant cells were tested in a luciferase assay after induction with murine IL-3 to establish inducibility of the luciferase reporter gene in these cells. Clones responsive to murine IL-3 (BAF3/LUC cells) were further expanded and used for introduction of human EPOR. For introduction of human EPO, BAF3/LUC cells maintained in Zeocine were electroporated in the presence of RcCMV/EPOR and selected in the presence of 200 μg/ml Geneticin (GIBCO/BRL) for 2 wk. The cultures were further selected in EPO (1 unit/ml) to obtain a pool of BAF3/LUC cells expressing the functional EPOR receptor.
Luciferase Assay.
Fifty thousand cells in 200 μl of RPMI 1640 containing 10% FBS were plated into each well of 96-well plates (Falcon). After 16–20 hr of incubation with cytokines or compounds, the luciferase activity was determined after addition of an equal volume of LucLite Reagent (Packard) and luminescence measured in a MLX Microtiterplate Luminometer (Dynatech).
Mitogenic Assay.
Mouse BAF3 cells expressing human EPOR, DA3 cells expressing human and mouse EPOR, parental BAF3 and DA3 cells, and human erythroleukemia cell lines F36 and TF-1 were plated at a density of 10,000–20,000 cells per well in 96-well microtiter plates in RPMI 1640 containing 10% FBS. After 40 hr of incubation with cytokines or compounds, 3H-thymidine (4 μCi/ml) was added to each well and allowed to incorporate into DNA for 4 hr. The cells were harvested onto a LKB Filtermat B by using a Tomtec Cell Harvester Mach II (Tomtec, Orange, CT). Filtermats were dried, sealed in counting bags with 23-ml scintillation mixture, and counted on a Wallac (Gaithersburg, MD) 1205 Betaplate counter.
Hematopoietic Colony Assays.
Colony assays were set up by using human CD34+ mononuclear cells isolated from healthy donors. One thousand to two thousand cells were mixed with 1 ml of methylcellulose culture medium containing FBS (StemCell Technologies, Vancouver), 0.4% DMSO, a mixture of growth factors containing granulocyte–macrophage colony stimulating factor (GM-CSF), IL-3, granulocyte colony stimulating factor (G-CSF), and stem cell factor (HC-4535-StemCell Technologies, Vancouver), and EPO (Epoetin Alfa, Amgen Biologicals) or compounds. After thorough mixing, the suspension was plated in 35-mm gridded plates (Nunc) and incubated at 37°C in a humidity controlled CO2 incubator. Colony-forming units (CFU)–erythroid containing >50 hemoglobinized cells and myeloid colonies (CFU-GM) containing >50 cells were counted on day 12–14. Mixed colonies, containing both erythroid and myeloid cells, were counted on day 14–16. All assays were performed at Poietic Technologies, Gaithersburg, MD.
Results and Discussion
Compound 5, an Antagonist of EPO-Binding to EPOR, also Induces Dimerization of the EPOR.
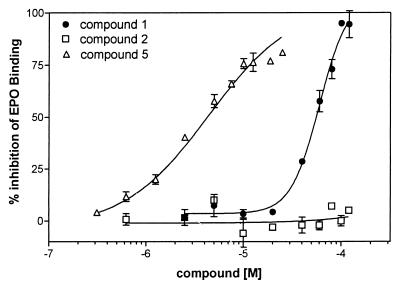
To test the hypothesis that a multimeric form of an EPOR antagonist can function as an EPO mimetic to dimerize and activate the EPOR, we screened the in-house chemical collection for inhibitors of EPO binding to the EPOR using the extracellular domain of the human EPOR as rEBP. This screening resulted in identification of N-3-[2-(4-biphenyl)-6-chloro-5-methyl]indolyl-acetyl-l-lysine methyl ester (compound 1). Compound 1 inhibited binding of 125I-EPO to the rEBP with an IC50 of 59.5 (+/− 1.1) μM (Fig. 2). Compound 5, a precursor in the synthesis of compound 1, consists of eight copies of compound 1 attached to a polyamidoamino-octa-4-hydroxymethylbenzamide support via a chemical linker (compound 2). When tested in the EPOR binding assay, compound 5 also inhibited binding of 125I-EPO to the rEBP with an IC50 of 4.4 +/− 1.9 μM (Fig. 2). Compound 2, the unconjugated dendrimer used to crosslink compound 1, did not affect binding of 125I-EPO to the rEBP in this assay. The ability of compound 5 to inhibit binding of 125I-EPO to the rEBP suggests that compound 1, even when attached to an inactive central core, is able to interact with rEBP.
Figure 2.
Inhibition of 125I-EPO binding to rEBP. Dose-response curve for inhibition of 125I-EPO binding by compounds 1, 2, and 5. Competition binding experiments were performed in a volume of 50 μl containing 5 nM 125I-EPO in the presence or absence of compounds and allowed to proceed for at least 16 hr at 4°C, as described in Materials and Methods. Nonspecific EPO binding was determined by performing the experiments in the presence of 1 μM unlabeled EPO and was found to be less than 10% (under these conditions, we routinely obtained a total of 3,000–5,000 counts per minute (cpm)/well with no compound and 200–400 cpm/well in the presence of 1 μM cold EPO). The data are expressed as a percentage of the EPO (specific cpm) bound in the presence of 5% DMSO but in the absence of any compound (which is considered as 100%). Each data point was analyzed in triplicate and is a mean (+/− SEM) of three independent experiments.
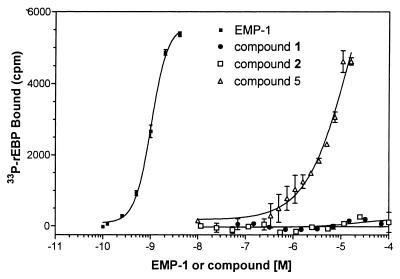
The biological activation of EPOR requires dimerization of EPOR on binding of EPO to two independent chains of EPOR. Thus, one of several ways for a small molecule to activate the EPOR is to induce dimerization of the EPOR. To determine whether compound 1 or 5 can dimerize EPOR in solution, we tested these compounds in an EPOR dimerization assay. This assay examined the interaction of a radiolabeled form of rEBP (33P-rEBP) in solution with a plate-immobilized rEBP. In the absence of any dimerizing agent in the assay, the labeled receptor fails to attach to the plate and is easily washed off during the washing step (at the end of the assay). However, in the presence of a compound that can interact with at least two chains of EPOR, the radiolabeled chain would be expected to remain bound to the plate, resulting in increased retention of 33P-rEBP in the assay. EMP-1, a 20-aa peptide, which has been shown to induce dimerization of this form of EPOR in solution (18), produces a dose-dependent increase in the retention of 33P-rEBP (Fig. 3). When tested in this assay, compound 5 produced a dose-dependent increase (EC50 15.9 +/− 3.3 μM) in retention of 33P-rEBP, whereas compounds 1 and 2 both failed to produce any such increases (Fig. 3). These data suggest that even though compounds 1 and 5 both compete for the binding of EPO to the rEBP, only compound 5 is able to interact with more than one chain of EPOR.
Figure 3.
Dimerization of soluble EPOR. 33P-rEBP was incubated with or without plate-immobilized rEBP (as in Fig. 2), in the absence or presence of compounds 1, 2, or 5 or EMP-1 for 16 hr at 4°C, as described in Materials and Methods. Data represent specific cpm (cpm with plate-immobilized EPOR minus cpm without plate-immobilized EPOR) observed with each of the compounds. Data are a mean of two experiments (+/− SEM) performed in triplicate. Under these conditions, mean cpm observed in wells with no compounds were 147 +/− 25).
Compound 5 Induces Cellular Transcription and Proliferation in Cells Expressing EPOR.
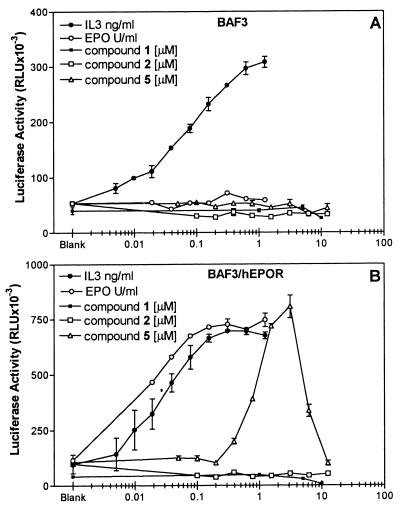
Binding of EPO to its cell-surface receptors results in the activation of cellular signaling pathways, including activation of janus kinase (JAK)2 and STAT5 (27–29). STAT5 is a transcription factor that translocates to the nucleus on activation and induces gene transcription (30). Luciferase reporter gene constructs containing synthetic promoters with STAT-binding sites have been used as markers for the activation of STATs in cells (31–33). To determine whether compound 5 could interact with the EPOR expressed in mammalian cells, induce receptor oligomerization as seen in vitro (Fig. 3), and activate the JAK-STAT signaling pathway, we tested this compound on BAF3/LUC cells. The BAF3/LUC cells have a stably integrated luciferase gene under the control of a STAT-binding site from the interferon regulatory factor 1 gene. This STAT-binding site has been shown to function in response to activation signals involving STATs 1–5 (25, 34–36). Because BAF3 cells do not normally express EPORs, they do not manifest any biological response when treated with EPO; however, they can acquire EPO responsiveness once the EPOR is expressed ectopically (37, 38). However, these cells do express IL-3 receptors and in response to IL-3 activate the JAK2 and STAT5 signaling pathway (39–41). As shown in Fig. 4, treatment with IL-3 increases the luciferase activity in both BAF3/LUC and BAF3/LUC/EPOR cells, demonstrating that these cells are competent in responding to receptor-dependent activation of Stat5. As expected, EPO-dependent increases in the luciferase activity were observed in the EPO-treated BAF3/LUC/EPOR cells but not in EPO-treated BAF3/LUC cells. When tested in these cells, compound 5 induced increases in the luciferase activity only in the BAF3/LUC/hEPOR cells with an EC50 of 1.25 μM. No such increases were observed in BAF3/LUC cells (Fig. 4). Moreover, neither compound 1 nor compound 2 induced any increases in the luciferase activity in either of these two cell lines. The decrease in the luciferase activity observed at higher concentrations of compound 5 may be caused by toxicity or by the engagement of EPOR in a 1:1 (EPOR/compound) complex, thus preventing the formation of EPOR dimers needed for the activation of the JAK-STAT pathway. Formation of this 1:1 receptor/ligand complex at high ligand concentration has been shown to inhibit the activation of growth hormone and EPORs by their respective ligands (17, 42).
Figure 4.
Induction of luciferase activity in BAF3/LUC and BAF3/LUC/EPOR cells. BAF3/LUC or BAF3/LUC/EPOR cells containing a stably integrated luciferase gene under the control of a STAT-binding element were treated with varying amounts of EPO, IL-3, or compounds for 16 hr. The luciferase activity was determined as described in Materials and Methods and is expressed in relative light units (RLU). (A) Luciferase activity in BAF3/LUC cells. (B) Luciferase activity in BAF3/LUC/EPOR cells, expressing human EPOR. Data are a mean (+/− SEM) of two to three independent experiments performed in triplicate. Value of x axis reflects the concentration of EPO, IL-3, or compounds in unit/ml, ng/ml, and μM, respectively. All assays were performed in 1% DMSO. Values shown above blank on graphs refer to luciferase activity observed in the untreated cultures in the presence of 1% DMSO.
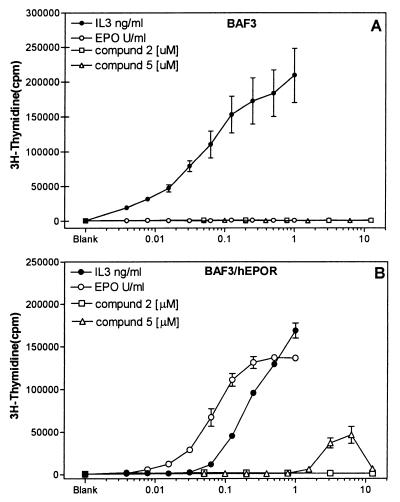
BAF3 cells are known to proliferate in the presence of IL-3 and a number of other cytokines, including EPO, provided that the receptors for these cytokines are expressed in these cells. The increased proliferation results in increased incorporation of nucleotides, such as a thymidine, into DNA, which can be measured by using 3H-thymidine. Much like what was observed for the luciferase reporter gene activation, IL-3 induced increases in the 3H-thymidine incorporation in both BAF3 and BAF3/hEPOR cells, but EPO induced these increases in BAF3/hEPOR cells only (Fig. 5). When tested in these cells, compound 5 induced 3H-thymidine incorporation in BAF3/hEPOR cells but not in parental BAF3 cells, similar to the response to EPO. However, compound 2 failed to show any activity in the assay (because of lack of activity in the EPOR dimerization and luciferase reporter gene assay, compound 1 was not evaluated in this or any subsequent assays), and again compound 5 showed a decreased activity at higher concentrations, which could be caused by nonspecific cellular toxicity or unproductive engagement of receptors (as suggested above). BAF3 cells require the presence of a cytokine such as IL-3 even for survival in cell culture, thus making it difficult to distinguish between these two possibilities. However, this phenomenon is not limited to BAF3 cells, because we have observed similar inhibition at high concentrations in DA3/hEPOR cells, another murine cell line, as well as in F36E and TF-1 cells, two cell lines of human origin known to respond to EPO via the endogenous EPOR (23, 24). Moreover, we have observed a compound 5-dependent inhibition of IL-3-induced thymidine incorporation in BAF3 cells lacking the EPOR (S.Q. and R.R., unpublished observation), which supports the notion that the toxicity of compound 5 is nonspecific and independent of the EPOR. More experiments will be needed to sort out this phenomenon. Regardless of the complexity in the interpretation of the data at higher doses of compound 5, these data still demonstrate that compound 5 interacts with the EPOR expressed on the cell surface, resulting in induction of EPO-like activities.
Figure 5.
Mitogenic responses in BAF3 and BAF3/hEPOR cells. BAF3 or BAF3/hEPOR cells were treated with different amounts of EPO, IL-3, or compounds for 48 hr. 3H-thymidine incorporation, as an indicator of cell proliferation, was measured after addition of 4 μCi/ml of 3H-thymidine during the last 4 hr of incubation, as described in Materials and Methods. (A) Amount of 3H-thymidine incorporated in BAF3 cells after different treatments. (B) Effect of same in BAF3 cells expressing human EPOR. Data represent mean (+/− SEM) of two to three experiments where each determination was made in triplicate. Value of x axis reflects the concentration of EPO, IL-3, or compounds in units/ml, ng/ml, and μM, respectively. All assays were performed in 1% DMSO. Values shown above blank on graphs refer to level of radioactivity incorporated in the untreated cultures in the presence of 1% DMSO.
Compound 5 Induces Differentiation of Human Progenitor Cells into Cells of Erythrocytic Lineage.
EPO induces differentiation of hematopoietic progenitor cells into mature erythrocytes in vivo. However, in tissue culture conditions, only a part of this process is recapitulated. Progenitor cells isolated from peripheral blood or bone marrow when cultured in vitro in semisolid media can grow into morphologically distinct colonies (each colony being a product of single progenitor cell), depending on which growth factor is present in the media. For example, it is well known that EPO, in the presence of stem cell factor and IL-3, induces the formation of colonies known as CFU-E (Colony Forming Unit–Erythroid), which are easily identified microscopically because of the hemoglobinization of cells in the colony. Cells isolated from human peripheral blood by using an anti-CD34 antibody (CD34+ cells) are more advanced progenitors capable of differentiation into erythrocytes in the presence of EPO and into granulocytes and macrophages in the presence of G-CSF and GM-CSF (43, 44). We used these cells to examine the ability of compound 5 to replace EPO in promoting differentiation of these progenitor cells into erythroid colonies in vitro. Cells plated in the presence of growth factors (stem cell factor, IL-3, GM-CSF, and G-CSF) alone did not develop into erythroid colonies, even though these were able to form colonies of GM progenitor cells (CFU-GM) indicating the viability of these isolated cells (Table 1). However, addition of EPO to the mixture of growth factors induced formation of erythroid colonies in a dose-dependent manner. Under the same conditions, compound 5 also induced erythroid colonies, whereas compound 2 did not induce erythroid colonies and did not inhibit the formation of GM colonies. Moreover, no morphological differences could be observed between the EPO-induced erythroid colonies or the compound-induced erythroid colonies. These data confirm that compound 5 is an EPO mimetic capable of inducing the proliferation and differentiation of human erythroid progenitor cells.
Table 1.
Induction of CFU-erythroid colonies by EPO and compound 5
| Treatment | Dose | Avg. no. colonies per treatment
|
|
|---|---|---|---|
| Erythroid colonies | GM colonies | ||
| None | — | 0 ± 0 | 86 ± 13 |
| DMSO | 0.4% | 0 ± 0 | 74 ± 2 |
| EPO | 100 mU/ml | 66 ± 1 | 87 ± 7 |
| DMSO + EPO | 100 mU/ml | 55 ± 2 | 73 ± 5 |
| 30 mU/ml | 48 ± 3 | 73 ± 5 | |
| 10 mU/ml | 20 ± 7 | 72 ± 13 | |
| 3 mU/ml | 2 ± 1 | 65 ± 5 | |
| Compound 2 | 2000 nM | 0 ± 0 | 65 ± 15 |
| 500 nM | 0 ± 0 | 66 ± 11 | |
| 125 nM | 0 ± 0 | 72 ± 11 | |
| Compound 5 | 2000 nM | 47 ± 2 | 65 ± 1 |
| 1000 nM | 43 ± 1 | 76 ± 1 | |
| 500 nM | 30 ± 2 | 75 ± 5 | |
| 250 nM | 12 ± 2 | 71 ± 1 | |
| 125 nM | 3 ± 1 | 75 ± 1 | |
Human CD34+ progenitor cells (1.5 × 103/plate) were plated in semisolid media to allow growth of erythroid colonies and GM colonies. Colonies of each type were counted after 12–15 days of seeding under each condition. Data are mean (SEM) of two independent experiments, where each determination was performed in triplicate.
In summary, we have reported on the identification and characterization of an EPOR antagonist, which when presented in a multivalent configuration induced biological responses similar to what is normally observed with EPO. Although the potency of the activities observed with this compound is only a fraction of what is expected of EPO, it does validate the concept that the EPOR, and by extension most cytokine receptors, can be ligated together in an active conformation by a nonpeptidyl molecule. The only requirement is that the small molecule must be able to interact with both chains of the receptor. This paper also lays out a basic strategy for identifying cytokine mimetics by converting an antagonist into an agonist.
Acknowledgments
We thank Drs. Alan D’Andrea, Jim Ihle, and K. Kitamura and the Rieken Cell Bank for cell lines used in these studies, and Poietic Technologies, Inc. for performing the hematopoietic colony assays. We also thank Steve Parent, Kristine Prendergast, and Bruce Bush for helpful discussions, Bruce Bush, David E. Moller, and Russell B. Lingham for comments on the manuscript, and Roy Smith and Bennett Shapiro for their encouragement and support.
Abbreviations
- EPO
erythropoietin
- 125I-EPO
(3-[125I] iodotyrosyl) EPO
- EPOR
EPO receptor
- rEBP
recombinant EPO-binding protein
- EMP-1
EPO mimetic peptide-1
- STAT
signal transducer and activator of transcription
- JAK
janus kinase
- CFU
colony-forming unit
- GM-CSF
granulocyte–macrophage colony stimulating factor
- G-CSF
granulocyte colony stimulating factor
- 33P-rEBP
radiolabeled form of rEBP
- cpm
counts per minute
- DMF
N,N-dimethylformamide
- SEC
size exclusion chromatography
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Koury M J, Bondurt M C. J Cell Physiol. 1988;137:65–74. doi: 10.1002/jcp.1041370108. [DOI] [PubMed] [Google Scholar]
- 2.Krantz S. Blood. 1991;77:419–434. [PubMed] [Google Scholar]
- 3.Koury S T, Bondurant M C, Koury M J. Blood. 1988;71:524–527. [PubMed] [Google Scholar]
- 4.Lacombe C D, De Silva J L. J Clin Invest. 1988;81:621–623. doi: 10.1172/JCI113363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jelkmann W. Physiol Rev. 1992;72:449–489. doi: 10.1152/physrev.1992.72.2.449. [DOI] [PubMed] [Google Scholar]
- 6.Maxwell P H, Ferguson D J P, Nicholls L G, Iredale J P, Pugh C W, Johnson M H, Ratcliffe P J. Kidney Int. 1997;51:393–401. doi: 10.1038/ki.1997.52. [DOI] [PubMed] [Google Scholar]
- 7.Valderrabano F. Kidney Int. 1996;50:1373–1391. doi: 10.1038/ki.1996.452. [DOI] [PubMed] [Google Scholar]
- 8.Spivak J L. Annu Rev Med. 1993;44:243–253. doi: 10.1146/annurev.me.44.020193.001331. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Gagnon R F, Richards G K, Brox A G. Nephron. 1996;72:654–661. doi: 10.1159/000188955. [DOI] [PubMed] [Google Scholar]
- 10.Brox A G, Zhang F, Guyda H, Gagnon R F. Kidney Int. 1996;50:937–943. doi: 10.1038/ki.1996.394. [DOI] [PubMed] [Google Scholar]
- 11.Wells J. Cell Biol. 1994;6:163–173. doi: 10.1016/0955-0674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 12.Heldin C-H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura A, Longmore G, Lodish H. Nature (London) 1990;348:647–649. doi: 10.1038/348647a0. [DOI] [PubMed] [Google Scholar]
- 14.Watowich S, Yoshimura A, Longmore G, Hilton D, Yoshimura Y, Lodish H. Proc Natl Acad Sci USA. 1992;89:2140–2144. doi: 10.1073/pnas.89.6.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watowich S S, Hilton D J, Lodish H F. Mol Cell Biol. 1994;14:3535–3549. doi: 10.1128/mcb.14.6.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longmore G, Pharr P, Lodish H. Mol Cell Biol. 1994;14:2266–2277. doi: 10.1128/mcb.14.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider H, Chaovapong W, Matthews D J, Karkaria C, Cass R T, Zhan H J, Boyle M, Lorenzini T, Elliott S G, Giebel L B. Blood. 1997;89:473–482. [PubMed] [Google Scholar]
- 18.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 19.Livnah O, Stura E A, Johnson D L, Middleton S A, Mulcahy L S, Wrighton N C, Dower W J, Jolliffe L K, Wilson I A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 20.Syed R S, Reid S W, Li C, Cheetham J C, Aoki K N, Liu B, Zhan H, Osslund T D, Chirino A J, Zhang J, et al. Nature (London) 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 21.Kim R M, Manna M, Hutchins S M, Griffin P R, Yates N A, Bernick A M, Chapman K T. Proc Natl Acad Sci USA. 1996;93:10012–10017. doi: 10.1073/pnas.93.19.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penninghton, M. W. & Dunn, B. M. (194) in Methods in Molecular Biology (Humana, Totowa, NJ), Vol. 35.
- 23.Chiba S, Takaku F, Tange T, Shibuya K, Misawa C, Sasaki K, Miyagawa K, Yazaki Y, Hirai H. Blood. 1991;78:2261–2268. [PubMed] [Google Scholar]
- 24.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao Y-F, Miyazono K, Urabe A, Takaku F. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 25.Rosenblum C I, Tota M, Cully D, Smith T, Collum R, Qureshi S, Hess J F, Phillips M S, Hey P J, Vongs A, et al. Endocrinology. 1996;137:5178–5181. doi: 10.1210/endo.137.11.8895396. [DOI] [PubMed] [Google Scholar]
- 26.Johnson D, Middleton S, McMahon F, Barbone F, Kroon D, Tsao E, Lee W, Mulcahy L, Jolliffe L. Protein Expression Purif. 1996;7:104–113. doi: 10.1006/prep.1996.0014. [DOI] [PubMed] [Google Scholar]
- 27.Gouilleux F, Pallard C, Dusanterfourt I, Wakao H, Haldosen L A, Norstedt G, Levy D, Groner B. EMBO J. 1995;14:2005–2013. doi: 10.1002/j.1460-2075.1995.tb07192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quelle F W, Wang D, Nosaka T, Thierfelder W E, Stravopodis D, Weinstein Y, Ihle J N. Mol Cell Biol. 1996;16:1622–1631. doi: 10.1128/mcb.16.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihle J N. Philos Trans R Soc London B. 1996;351:159–166. doi: 10.1098/rstb.1996.0012. [DOI] [PubMed] [Google Scholar]
- 30.Penta K, Sawyer S T. J Biol Chem. 1995;270:31282–31287. doi: 10.1074/jbc.270.52.31282. [DOI] [PubMed] [Google Scholar]
- 31.Wakao H, Gouilleux F, Groner B. EMBO J. 1994;13:2182–2191. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb P, Seidel H M, Haslam J, Milocco L, Kessler L V, Stein R B, Rosen J. Nucleic Acids Res. 1995;23:3283–3289. doi: 10.1093/nar/23.16.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidel H M, Milocco L H, Lamb P, Darnell J E, Stein R B, Rosen J. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman P, Kreider B, Azam M, Levy D, Wegenka U, Eilers A, Decker T, Horn F, Kashleva H, Ihle J, Schindler C. Immunity. 1994;1:457–468. doi: 10.1016/1074-7613(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 35.Schindler C. Receptor. 1995;5:51–62. [PubMed] [Google Scholar]
- 36.Darnell J E. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 37.Barber D L, Mason J M, Fukazawa T, Reedquist K A, Druker B J, Band H, D’Andrea A D. Blood. 1997;89:3166–3174. [PubMed] [Google Scholar]
- 38.Pless M, Norga K, Carroll M, Heim M H, Dandrea A D, Matheyprevot B. Blood. 1997;89:3175–3185. [PubMed] [Google Scholar]
- 39.Azam M, Erdjument-Bromage H, Kreider B L, Xia M, Quelle F, Basu R, Saris C, Tempst P, Ihle J N, Schindler C. EMBO J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland N G, Gilbert D J, Jenkins N A, Hara T, Miyajima A. EMBO J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 42.Fuh G, Cunningham B, Fukunaga R, Nagata S, Goeddel D, Wells J. Science. 1992;256:1677–1680. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- 43.Testa U, Fossati C, Samoggia P, Masciulli R, Mariani G, Hassan H J, Sposi N M, Guerriero R, Rosato V, Gabbianelli M, et al. Blood. 1996;88:3391–3406. [PubMed] [Google Scholar]
- 44.Alcorn M J, Richmond H L, Pearson C, Farrel E, Kyle B, Dunlop D J, Fitzsimons E, Steward W P, Pragnell I B, Franklin I M. J Clin Oncol. 1996;14:1839–1847. doi: 10.1200/JCO.1996.14.6.1839. [DOI] [PubMed] [Google Scholar]