Abstract
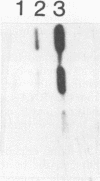
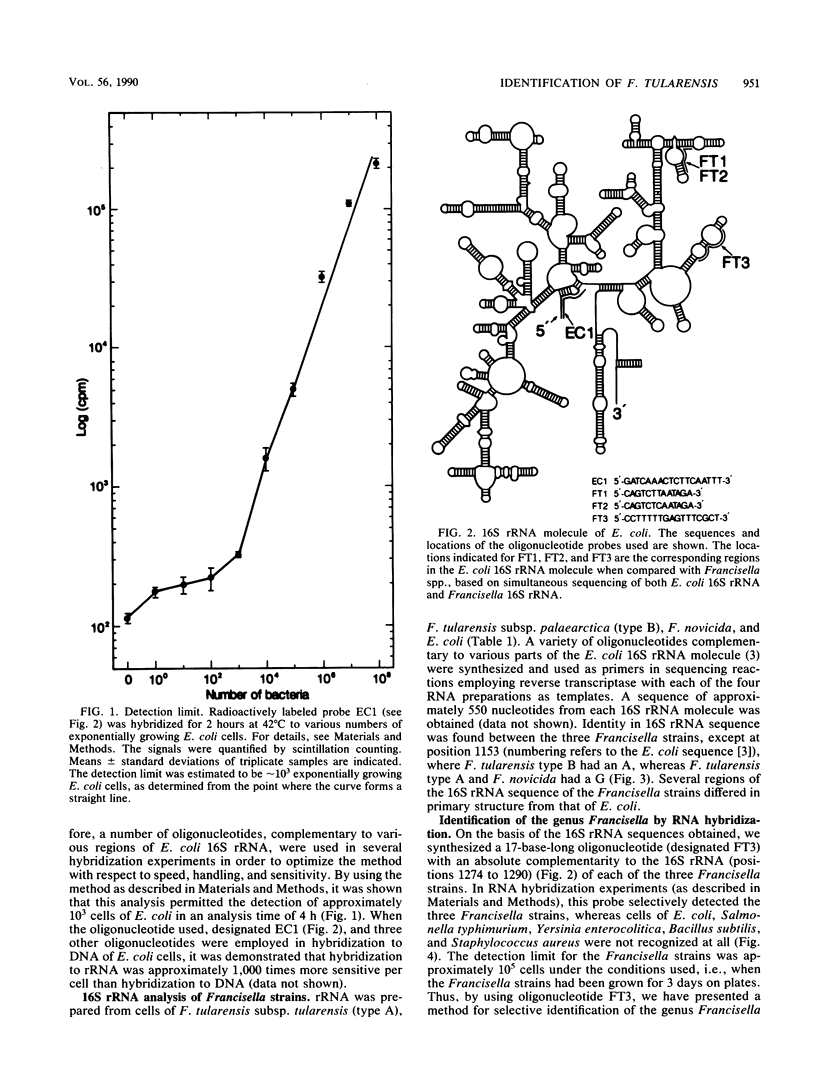
Tularemia is a zoonotic disease, occurring throughout the Northern Hemisphere. The causative agent, the bacterium Francisella tularensis, is represented by two main types. Type A is found in North America, whereas type B is mainly found in Asia and Europe and to a minor extent in North America. No routine technique for rapid diagnosis of tularemia has been generally applied. We have partially sequenced 16S rRNAs of two F. tularensis strains, as well as the closely related Francisella novicida. Of 550 nucleotides analyzed, only one difference in 16S rRNA primary sequence was found. This 16S rRNA analysis enabled the construction of oligonucleotides to be used as genus- and type-specific probes. Such probes were utilized for the establishment of a method for rapid and selective detection of the organism. This method allowed identification of Francisella spp. at the level of genus and also discrimination of type A and type B strains of F. tularensis. The analysis also permitted the detection of F. tularensis in spleen tissue from mice infected with the bacterium. The results presented will enable studies on the epizootiology and epidemiology of Francisella spp.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARLISLE H. N., HINCHLIFFE V., SASLAW S. Immunodiffusion studies with Pasteurella tularensis antigen-rabbit antibody systems. J Immunol. 1962 Nov;89:638–644. [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Fellous M., Nir U., Wallach D., Merlin G., Rubinstein M., Revel M. Interferon-dependent induction of mRNA for the major histocompatibility antigens in human fibroblasts and lymphoblastoid cells. Proc Natl Acad Sci U S A. 1982 May;79(10):3082–3086. doi: 10.1073/pnas.79.10.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman M., Lindgren L., Häggström B., Jaurin B. Transcriptional induction of Streptomyces cacaoi beta-lactamase by a beta-lactam compound. Mol Microbiol. 1989 Oct;3(10):1425–1432. doi: 10.1111/j.1365-2958.1989.tb00125.x. [DOI] [PubMed] [Google Scholar]
- Göbel U. B., Geiser A., Stanbridge E. J. Oligonucleotide probes complementary to variable regions of ribosomal RNA discriminate between Mycoplasma species. J Gen Microbiol. 1987 Jul;133(7):1969–1974. doi: 10.1099/00221287-133-7-1969. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Ivanov I., Gigova L. RNA colony hybridization method. Gene. 1986;46(2-3):287–290. doi: 10.1016/0378-1119(86)90413-0. [DOI] [PubMed] [Google Scholar]
- Jantzen E., Berdal B. P., Omland T. Cellular fatty acid composition of Francisella tularensis. J Clin Microbiol. 1979 Dec;10(6):928–930. doi: 10.1128/jcm.10.6.928-930.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Cohen S. N. Streptomyces lividans RNA polymerase recognizes and uses Escherichia coli transcriptional signals. Gene. 1984 Apr;28(1):83–91. doi: 10.1016/0378-1119(84)90090-8. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARCHETTE N. J., NICHOLES P. S. Virulence and citrulline ureidase activity of Pasteurella tularensis. J Bacteriol. 1961 Jul;82:26–32. doi: 10.1128/jb.82.1.26-32.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols P. D., Mayberry W. R., Antworth C. P., White D. C. Determination of monounsaturated double-bond position and geometry in the cellular fatty acids of the pathogenic bacterium Francisella tularensis. J Clin Microbiol. 1985 May;21(5):738–740. doi: 10.1128/jcm.21.5.738-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLSUFIEV N. G., EMELYANOVA O. S., DUNAYEVA T. N. Comparative study of strains of B. tularense in the old and new world and their taxonomy. J Hyg Epidemiol Microbiol Immunol. 1959;3:138–149. [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehnstam A. S., Norqvist A., Wolf-Watz H., Hagström A. Identification of Vibrio anguillarum in fish by using partial 16S rRNA sequences and a specific 16S rRNA oligonucleotide probe. Appl Environ Microbiol. 1989 Aug;55(8):1907–1910. doi: 10.1128/aem.55.8.1907-1910.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyeryar F. J., Lawton W. D. Factors Affecting Transformation of Pasteurella novicida. J Bacteriol. 1970 Dec;104(3):1312–1317. doi: 10.1128/jb.104.3.1312-1317.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tärnvik A., Löfgren S., Ohlund L., Sandström G. Detection of antigen in urine of a patient with tularemia. Eur J Clin Microbiol. 1987 Jun;6(3):318–319. doi: 10.1007/BF02017625. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]