Abstract
The bphC and bphD genes of Pseudomonas putida involved in the catabolism of polychlorinated biphenyls or biphenyl were identified, localized, and studied for expression in Escherichia coli. This was achieved by cloning a 2.4-kilobase (kb) DNA fragment of recombinant cosmid pOH101 into HindIII site of pUC plasmids downstream of a lacZ promoter and measuring the enzyme activities of 3-phenylcatechol dioxygenase (3-PDase; a product of bphC) and the meta-cleavage product 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (a product of bphD). The amount of 3-PDase produced in E. coli was about 20 times higher than that of the enzyme produced by the parent, P. putida. Determination of expression of the bphC and bphD genes through their own promoter sequences or by using the lacZ promoter of pUC plasmids was done by cloning the DNA that encodes bphC and bphD genes in a HindIII site of a promoter selection vector (pKK232-8) upstream of the gene for chloramphenicol acetyltransferase (CAT). The recombinant plasmid (pAW787) constructed by inserting the 2.4-kb DNA in pKK232-8 expressed both 3-PDase and CAT activities. Another hybrid construct (pAW786) in which the DNA insert was cloned in the opposite orientation lacked CAT activity but produced normal amounts of 3-PDase activity. On the basis of these results, we suggest that the bphC and bphD genes were expressed by using promoter sequences that are independent of the promoter that expresses CAT activity in E. coli. The locations of the bphC and bphD genes were determined by insertional inactivation of the open reading frames of structural genes bphC and bphD by Tn5 mutagenesis.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed M., Focht D. D. Degradation of polychlorinated biphenyls by two species of Achromobacter. Can J Microbiol. 1973 Jan;19(1):47–52. doi: 10.1139/m73-007. [DOI] [PubMed] [Google Scholar]
- Barton M. R., Crawford R. L. Novel biotransformations of 4-chlorobiphenyl by a Pseudomonas sp. Appl Environ Microbiol. 1988 Feb;54(2):594–595. doi: 10.1128/aem.54.2.594-595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard D. L., Haberl M. L., May R. J., Brennan M. J. Evidence for novel mechanisms of polychlorinated biphenyl metabolism in Alcaligenes eutrophus H850. Appl Environ Microbiol. 1987 May;53(5):1103–1112. doi: 10.1128/aem.53.5.1103-1112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984 Feb;27(2):151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Hayase N., Taira K., Tomizuka N. Molecular relationship of chromosomal genes encoding biphenyl/polychlorinated biphenyl catabolism: some soil bacteria possess a highly conserved bph operon. J Bacteriol. 1989 Oct;171(10):5467–5472. doi: 10.1128/jb.171.10.5467-5472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
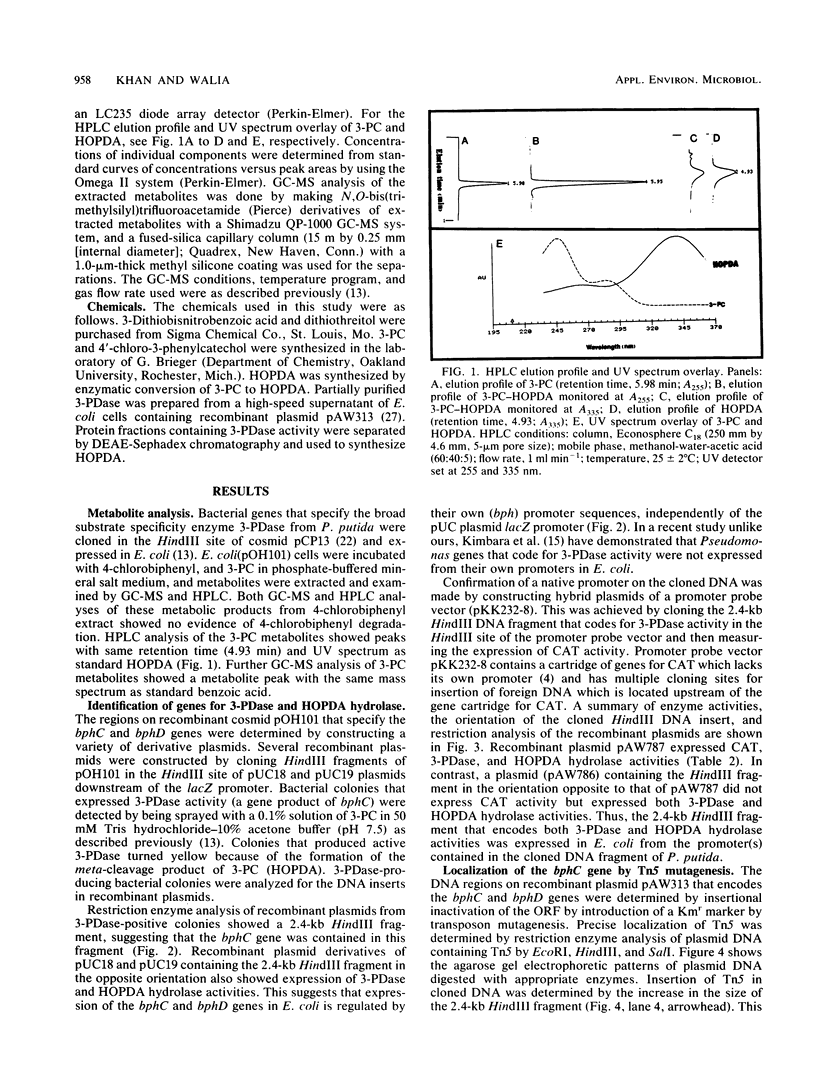
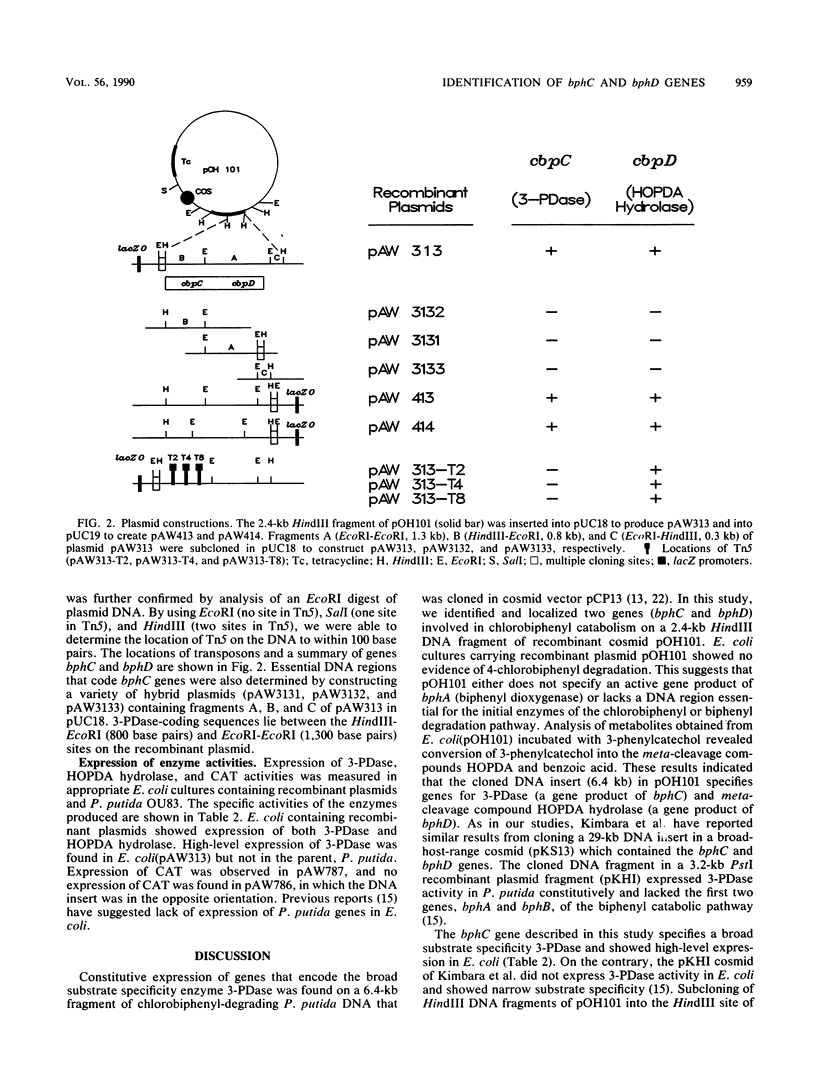
- Furukawa K., Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986 May;166(2):392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., You I. S., Chatterjee D. K., Chakrabarty A. M. Microbial degradation of halogenated compounds. Science. 1985 Apr 12;228(4696):135–142. doi: 10.1126/science.228.4696.135. [DOI] [PubMed] [Google Scholar]
- Ghosal D., You I. S., Gunsalus I. C. Nucleotide sequence and expression of gene nahH of plasmid NAH7 and homology with gene xylE of TOL pWWO. Gene. 1987;55(1):19–28. doi: 10.1016/0378-1119(87)90244-7. [DOI] [PubMed] [Google Scholar]
- Jones G. R. Polychlorinated biphenyls: where do we stand now? Lancet. 1989 Sep 30;2(8666):791–794. doi: 10.1016/s0140-6736(89)90844-1. [DOI] [PubMed] [Google Scholar]
- Khan A., Tewari R., Walia S. Molecular cloning of 3-phenylcatechol dioxygenase involved in the catabolic pathway of chlorinated biphenyl from Pseudomonas putida and its expression in Escherichia coli. Appl Environ Microbiol. 1988 Nov;54(11):2664–2671. doi: 10.1128/aem.54.11.2664-2671.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Walia S. Cloning of bacterial genes specifying degradation of 4-chlorobiphenyl from Pseudomonas putida OU83. Appl Environ Microbiol. 1989 Apr;55(4):798–805. doi: 10.1128/aem.55.4.798-805.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
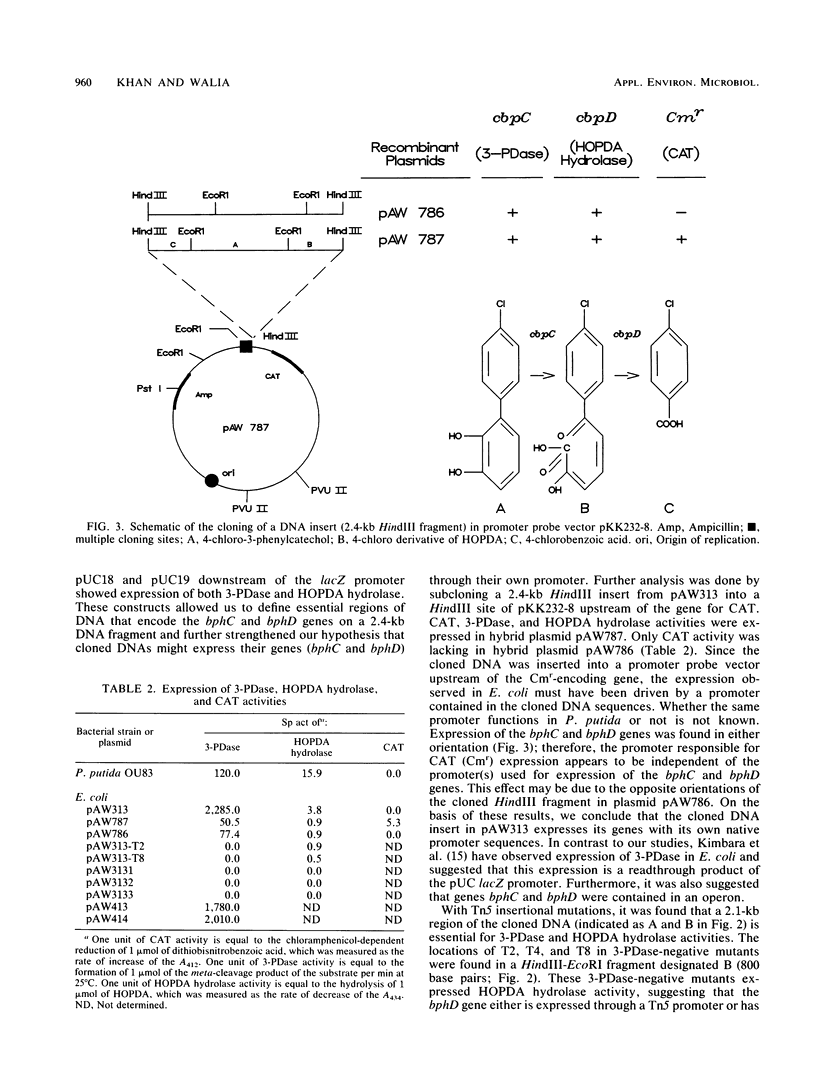
- Kimbara K., Hashimoto T., Fukuda M., Koana T., Takagi M., Oishi M., Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989 May;171(5):2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Mondello F. J. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol. 1989 Mar;171(3):1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangodkar U. M., Chapman P. J., Chakrabarty A. M. Cloning, physical mapping and expression of chromosomal genes specifying degradation of the herbicide 2,4,5-T by Pseudomonas cepacia AC1100. Gene. 1988 Nov 30;71(2):267–277. doi: 10.1016/0378-1119(88)90043-1. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Taira K., Hayase N., Arimura N., Yamashita S., Miyazaki T., Furukawa K. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene from the PCB-degrading strain of Pseudomonas paucimobilis Q1. Biochemistry. 1988 May 31;27(11):3990–3996. doi: 10.1021/bi00411a015. [DOI] [PubMed] [Google Scholar]
- Walia S. K., Duckworth D. H. The relationship of the delta transfer factor and KColIb to the ColIb plasmid. J Gen Microbiol. 1986 Dec;132(12):3261–3268. doi: 10.1099/00221287-132-12-3261. [DOI] [PubMed] [Google Scholar]
- Walia S., Khan A., Rosenthal N. Construction and applications of DNA probes for detection of polychlorinated biphenyl-degrading genotypes in toxic organic-contaminated soil environments. Appl Environ Microbiol. 1990 Jan;56(1):254–259. doi: 10.1128/aem.56.1.254-259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]