Abstract
A method for direct detection of Salmonella spp. in water was developed by using a commercially available DNA probe. Particulate DNA was extracted from 500- to 1,500-ml water samples collected from New York Harbor and Chesapeake Bay and used as a substrate for a salmonella-specific DNA probe in dot blot assays. The method detected salmonellae in water samples from 12 of 16 sites, including 6 sites where salmonellae could not be cultured. The specificity of the probe was evaluated, and cross-hybridization, although negligible, was used to set detection limits for the assay. Salmonella DNA bound the probe quantitatively, and from these results Salmonella DNA in the total particulate DNA in environmental samples could be estimated. The data obtained in this study indicate that Salmonella spp. often are not detected in water samples by culture methods, even when they are present in significant numbers.
Full text
PDF
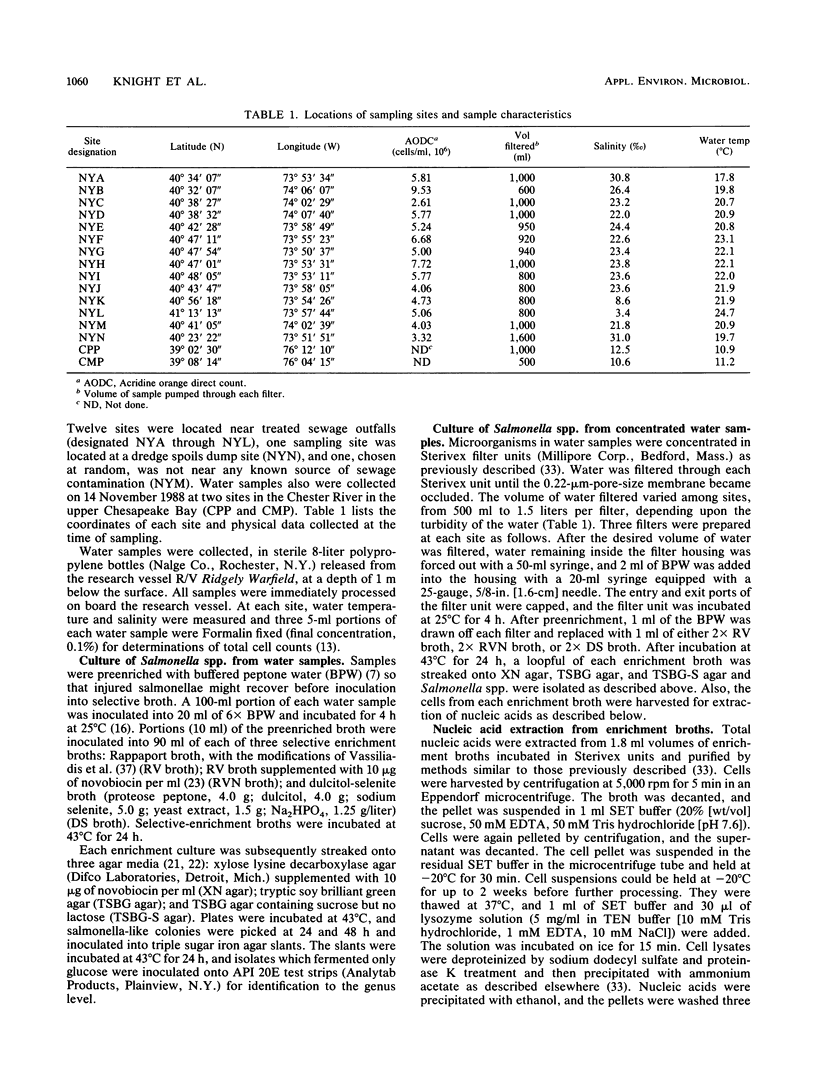

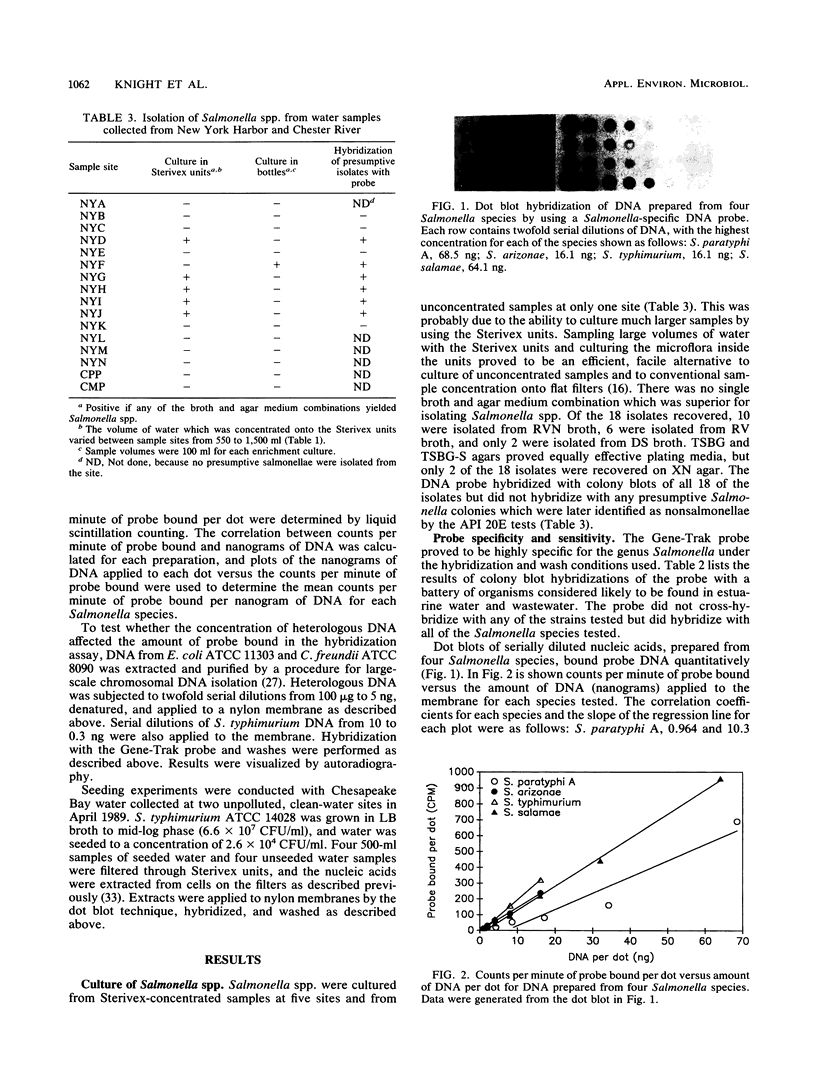




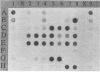
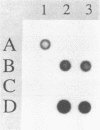
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcaide E., Martinez J. P., Garay E. Comparative study on Salmonella isolation from sewage-contaminated natural waters. J Appl Bacteriol. 1984 Jun;56(3):365–371. doi: 10.1111/j.1365-2672.1984.tb01363.x. [DOI] [PubMed] [Google Scholar]
- Anderson I. C., Rhodes M., Kator H. Sublethal stress in Escherichia coli: a function of salinity. Appl Environ Microbiol. 1979 Dec;38(6):1147–1152. doi: 10.1128/aem.38.6.1147-1152.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. M., Hartman P. A. Direct immunoassay for detection of salmonellae in foods and feeds. Appl Environ Microbiol. 1985 May;49(5):1124–1127. doi: 10.1128/aem.49.5.1124-1127.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird-Parker A. C., Boothroyd M., Jones E. The effect of water activity on the heat resistance of heat sensitive and heat resistant strains of salmonellae. J Appl Bacteriol. 1970 Sep;33(3):515–522. doi: 10.1111/j.1365-2672.1970.tb02228.x. [DOI] [PubMed] [Google Scholar]
- Bissonnette G. K., Jezeski J. J., McFeters G. A., Stuart D. G. Influence of environmental stress on enumeration of indicator bacteria from natural waters. Appl Microbiol. 1975 Feb;29(2):186–194. doi: 10.1128/am.29.2.186-194.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel W., Kampelmacher E. H. Comparative studies on the isolation of "sublethally injured" salmonellae in nine European laboratories. Bull World Health Organ. 1973;48(2):167–174. [PMC free article] [PubMed] [Google Scholar]
- Fitts R., Diamond M., Hamilton C., Neri M. DNA-DNA hybridization assay for detection of Salmonella spp. in foods. Appl Environ Microbiol. 1983 Nov;46(5):1146–1151. doi: 10.1128/aem.46.5.1146-1151.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopo J. M., Melis R., Filipska E., Meneveri R., Filipski J. Development of a Salmonella-specific biotinylated DNA probe for rapid routine identification of Salmonella. Mol Cell Probes. 1988 Dec;2(4):271–279. doi: 10.1016/0890-8508(88)90011-4. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. A., Ness G. E., Blake N. J. Relationship among fecal coliforms, Escherichia coli, and Salmonella spp. in shellfish. Appl Environ Microbiol. 1983 Jan;45(1):122–126. doi: 10.1128/aem.45.1.122-126.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussong D., Enkiri N. K., Burge W. D. Modified agar medium for detecting environmental salmonellae by the most-probable-number method. Appl Environ Microbiol. 1984 Nov;48(5):1026–1030. doi: 10.1128/aem.48.5.1026-1030.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Sayler G. S., Baldini M. M., Colwell R. R. Ambient-temperature primary nonselective enrichment for isolation of Salmonella spp. from an estuarine environment. Appl Environ Microbiol. 1977 Apr;33(4):829–835. doi: 10.1128/aem.33.4.829-835.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas R. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid. 1983 Nov;10(3):296–298. doi: 10.1016/0147-619x(83)90045-8. [DOI] [PubMed] [Google Scholar]
- Moats W. A. Comparison of four agar plating media with and without added novobiocin for isolation of salmonellae from beef and deboned poultry meat. Appl Environ Microbiol. 1978 Nov;36(5):747–751. doi: 10.1128/aem.36.5.747-751.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moats W. A., Kinner J. A. Observations on brilliant green agar with H2S indicator. Appl Environ Microbiol. 1976 Mar;31(3):380–384. doi: 10.1128/aem.31.3.380-384.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriñigo M. A., Borrego J. J., Romero P. Comparative study of different methods for detection and enumeration of Salmonella spp. in natural waters. J Appl Bacteriol. 1986 Aug;61(2):169–176. doi: 10.1111/j.1365-2672.1986.tb04272.x. [DOI] [PubMed] [Google Scholar]
- Moriñigo M. A., Martinez-Manzanares E., Muñoz A., Cornax R., Romero P., Borrego J. J. Evaluation of different plating media used in the isolation of salmonellas from environmental samples. J Appl Bacteriol. 1989 Apr;66(4):353–360. doi: 10.1111/j.1365-2672.1989.tb02488.x. [DOI] [PubMed] [Google Scholar]
- Rhodes M. W., Anderson I. C., Kator H. I. In situ development of sublethal stress in Escherichia coli: effects on enumeration. Appl Environ Microbiol. 1983 Jun;45(6):1870–1876. doi: 10.1128/aem.45.6.1870-1876.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Grimes D. J., Colwell R. R. Viable but nonrecoverable stage of Salmonella enteritidis in aquatic systems. Can J Microbiol. 1984 Mar;30(3):334–338. doi: 10.1139/m84-049. [DOI] [PubMed] [Google Scholar]
- Rubin F. A., Kopecko D. J., Noon K. F., Baron L. S. Development of a DNA probe to detect Salmonella typhi. J Clin Microbiol. 1985 Oct;22(4):600–605. doi: 10.1128/jcm.22.4.600-605.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. C., Knight I. T., Straube W. L., Colwell R. R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989 Mar;55(3):548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason B. M., Dodd D. J., Cherry W. B. Increased recovery of salmonellae from environmental samples enriched with buffered peptone water. Appl Environ Microbiol. 1977 Sep;34(3):270–273. doi: 10.1128/aem.34.3.270-273.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins L. S., Troup N., Labigne-Roussel A., Cohen M. L. Cloned, random chromosomal sequences as probes to identify Salmonella species. J Infect Dis. 1986 Jul;154(1):156–162. doi: 10.1093/infdis/154.1.156. [DOI] [PubMed] [Google Scholar]
- Vassiliadis P., Patéraki E., Papaïconomou N., Papadakis J. A., Trichopoulos D. Noueau procédé d'enrichissement de Salmonella. Ann Microbiol (Paris) 1976 Aug-Sep;127B(2):195–200. [PubMed] [Google Scholar]