Abstract
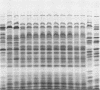
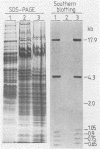
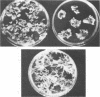
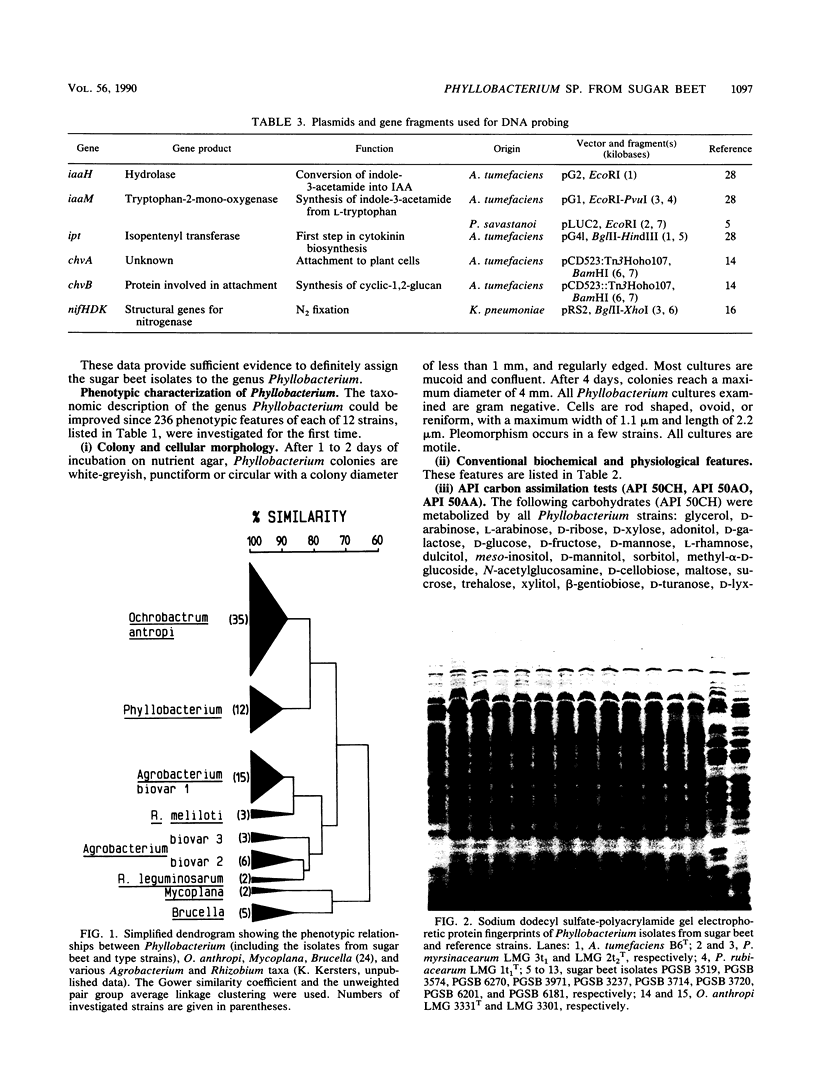
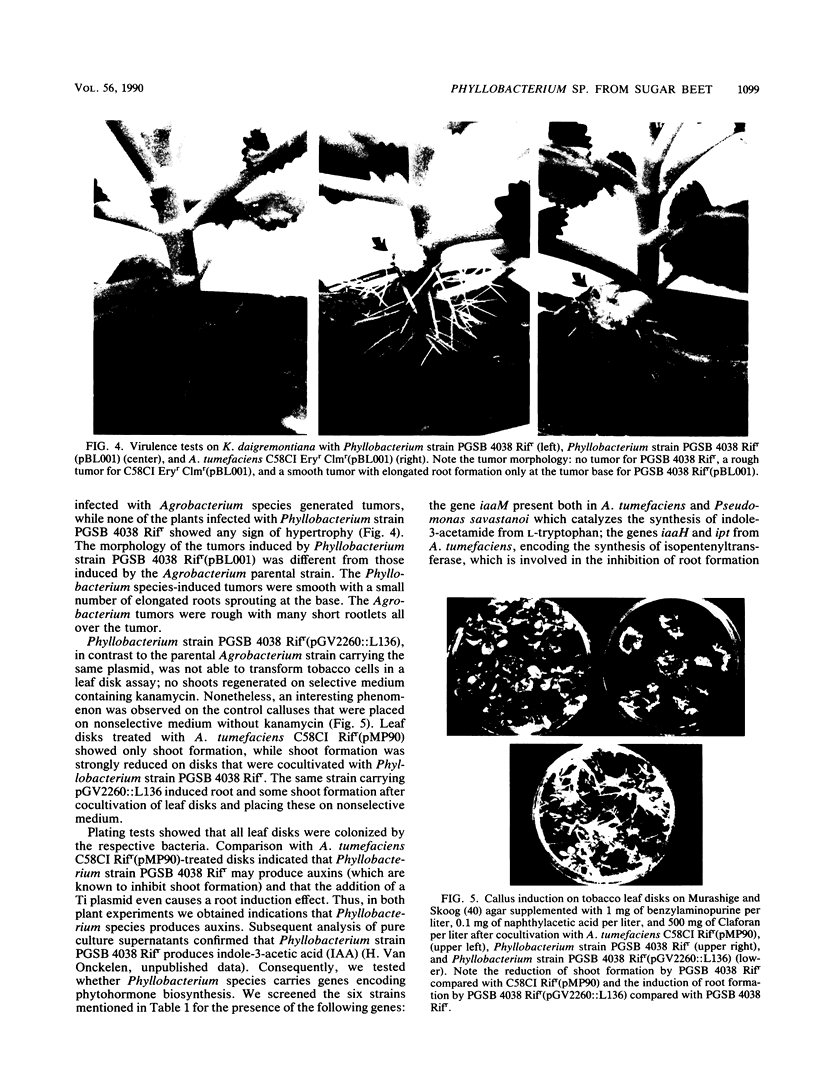
The second most abundant bacterium on the root surface of young sugar beet plants was identified as a Phyllobacterium sp. (Rhizobiaceae) based on a comparison of the results of 39 conventional identification tests, 167 API tests, 30 antibiotic susceptibility tests, and sodium dodecyl sulfate-polyacrylamide gel electrophoretic fingerprints of total cellular proteins with type strains of Phyllobacterium myrsinacearum and Phyllobacterium rubiacearum. It was found on 198 of 1,100 investigated plants between the 2nd and 10th leaf stage on three different fields in Belgium and one field in Spain. Densities ranged from 2 × 104 to 2 × 108 CFU/g of root. Five isolates exerted a broad-spectrum in vitro antifungal activity. DNA-DNA hybridizations showed that Phyllobacterium sp. does not contain DNA sequences that are homologous with the attachment genes chvA, chvB, the transferred-DNA (T-DNA) hormone genes iaaH and ipt from Agrobacterium tumefaciens, iaaM from A. tumefaciens and Pseudomonas savastanoi, or the nitrogenase genes nifHDK from Klebsiella pneumoniae. Phyllobacterium sp. produces indolylacetic acid in in vitro cultures and induces auxinlike effects when cocultivated with callus tissue of tobacco. When Phyllobacterium sp. was transformed with a Ti plasmid derivative, it gained the capacity to induce tumors on Kalanchoe daigremontiana. The potential role of Phyllobacterium sp. in this newly recognized niche is discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Block M. D., Botterman J., Vandewiele M., Dockx J., Thoen C., Gosselé V., Movva N. R., Thompson C., Montagu M. V., Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987 Sep;6(9):2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Kosuge T. Cloning characterization of iaaM, a virulence determinant of Pseudomonas savastanoi. J Bacteriol. 1982 Jan;149(1):40–46. doi: 10.1128/jb.149.1.40-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblaere R., Bytebier B., De Greve H., Deboeck F., Schell J., Van Montagu M., Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985 Jul 11;13(13):4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker A., De Wilde M., De Vos G., De Vos R., Van Montagu M., Schell J. Molecular cloning of overlapping segments of the nopaline Ti-plasmid pTiC58 as a means to restriction endonuclease mapping. Plasmid. 1980 Mar;3(2):193–211. doi: 10.1016/0147-619x(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Dhaese P., De Greve H., Decraemer H., Schell J., Van Montagu M. Rapid mapping of transposon insertion and deletion mutations in the large Ti-plasmids of Agrobacterium tumefaciens. Nucleic Acids Res. 1979 Dec 11;7(7):1837–1849. doi: 10.1093/nar/7.7.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas C. J., Staneloni R. J., Rubin R. A., Nester E. W. Identification and genetic analysis of an Agrobacterium tumefaciens chromosomal virulence region. J Bacteriol. 1985 Mar;161(3):850–860. doi: 10.1128/jb.161.3.850-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmerich C., Dreyfus B. L., Reysset G., Aubert J. P. Genetic analysis of nitrogen fixation in a tropical fast-growing Rhizobium. EMBO J. 1982;1(4):499–503. doi: 10.1002/j.1460-2075.1982.tb01197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett W. F., Osman S. F., Dunn M. F. Auxin production by plant-pathogenic pseudomonads and xanthomonads. Appl Environ Microbiol. 1987 Aug;53(8):1839–1845. doi: 10.1128/aem.53.8.1839-1845.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R. M., Hauptli H., Crossway A., Knauf V. C. Gene transfer in crop improvement. Science. 1987 Apr 3;236(4797):48–54. doi: 10.1126/science.236.4797.48. [DOI] [PubMed] [Google Scholar]
- HUGH R., LEIFSON E. The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram negative bacteria. J Bacteriol. 1953 Jul;66(1):24–26. doi: 10.1128/jb.66.1.24-26.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman L. M., Van Montagu M. C., Depicker A. G. Isolation of tobacco DNA segments with plant promoter activity. Mol Cell Biol. 1986 Dec;6(12):4486–4492. doi: 10.1128/mcb.6.12.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOVACS N. Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature. 1956 Sep 29;178(4535):703–703. doi: 10.1038/178703a0. [DOI] [PubMed] [Google Scholar]
- Lambert B., Leyns F., Van Rooyen L., Gosselé F., Papon Y., Swings J. Rhizobacteria of maize and their antifungal activities. Appl Environ Microbiol. 1987 Aug;53(8):1866–1871. doi: 10.1128/aem.53.8.1866-1871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse A. G. Characterization of nonattaching mutants of Agrobacterium tumefaciens. J Bacteriol. 1987 Jan;169(1):313–323. doi: 10.1128/jb.169.1.313-323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MØLLER V. Simplified tests for some amino acid decarboxylases and for the arginine dihydrolase system. Acta Pathol Microbiol Scand. 1955;36(2):158–172. doi: 10.1111/j.1699-0463.1955.tb04583.x. [DOI] [PubMed] [Google Scholar]
- PATON A. M. An improved method for preparing pectate gels. Nature. 1959 Jun 27;183:1812–1813. doi: 10.1038/1831812b0. [DOI] [PubMed] [Google Scholar]
- RHODES M. E. The cytology of Pseudomonas spp. as revealed by a silver-plating staining method. J Gen Microbiol. 1958 Jun;18(3):639–648. doi: 10.1099/00221287-18-3-639. [DOI] [PubMed] [Google Scholar]
- Robertson J. L., Holliday T., Matthysse A. G. Mapping of Agrobacterium tumefaciens chromosomal genes affecting cellulose synthesis and bacterial attachment to host cells. J Bacteriol. 1988 Mar;170(3):1408–1411. doi: 10.1128/jb.170.3.1408-1411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIERRA G. A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek. 1957;23(1):15–22. doi: 10.1007/BF02545855. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]