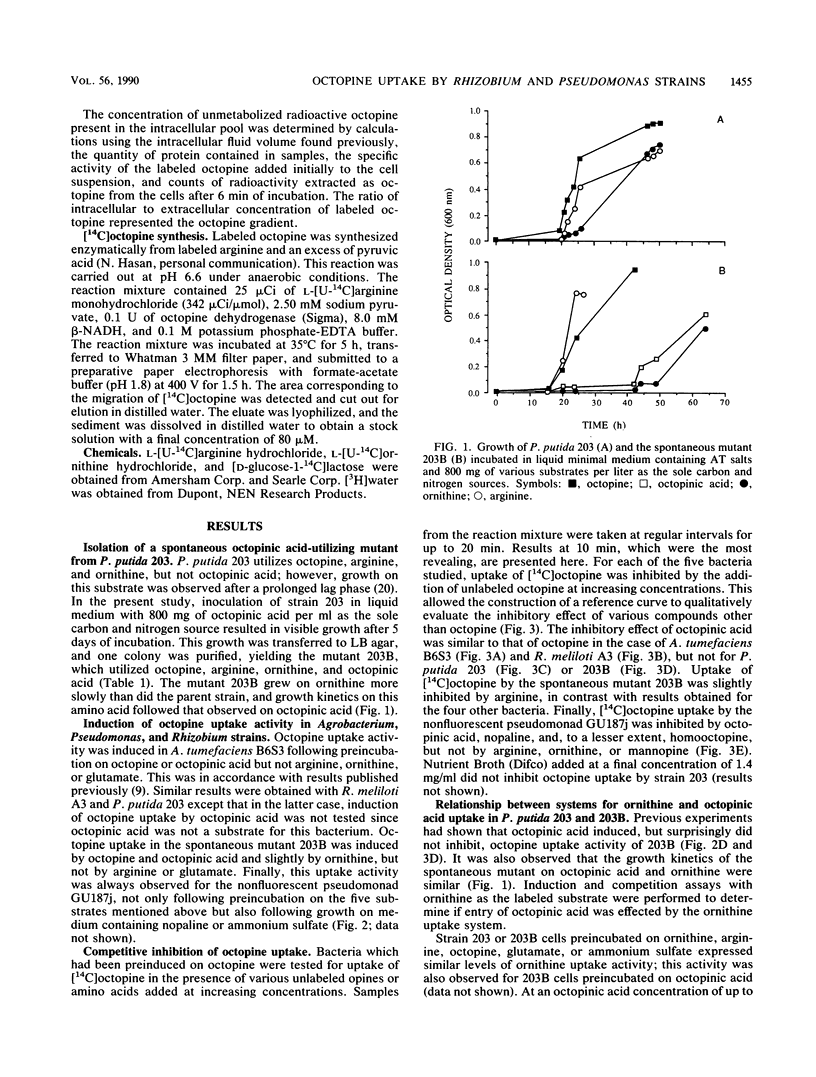
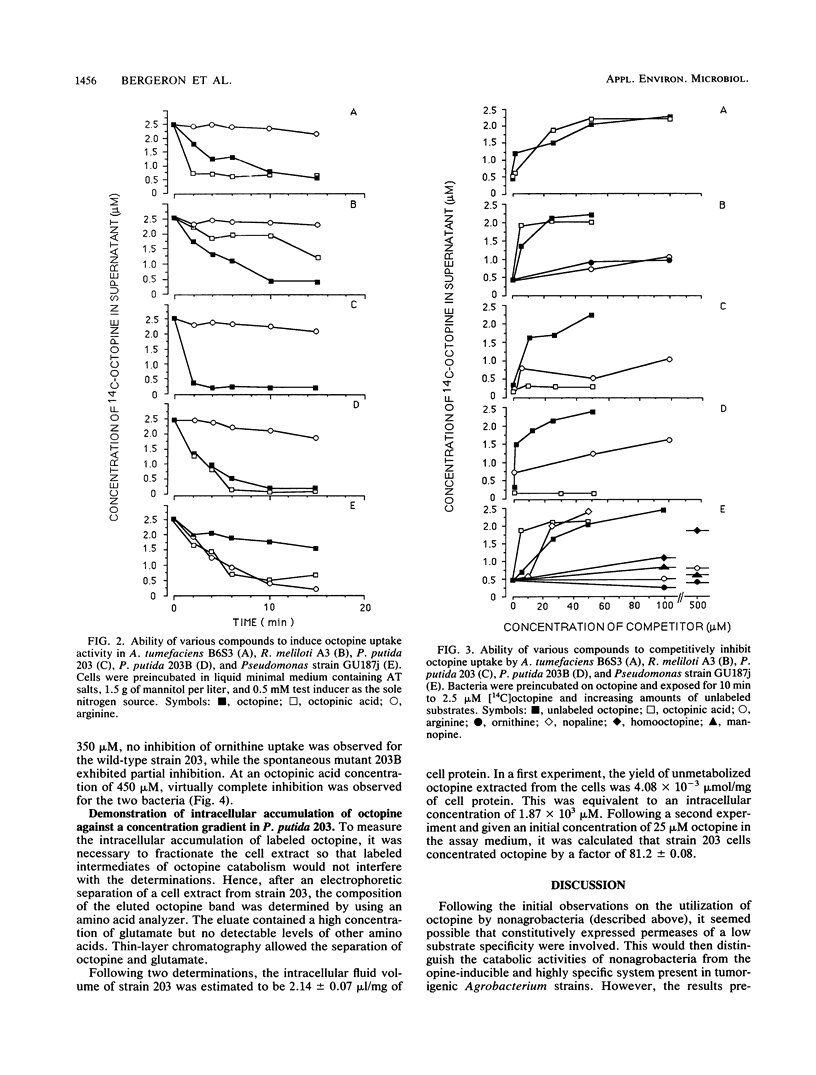
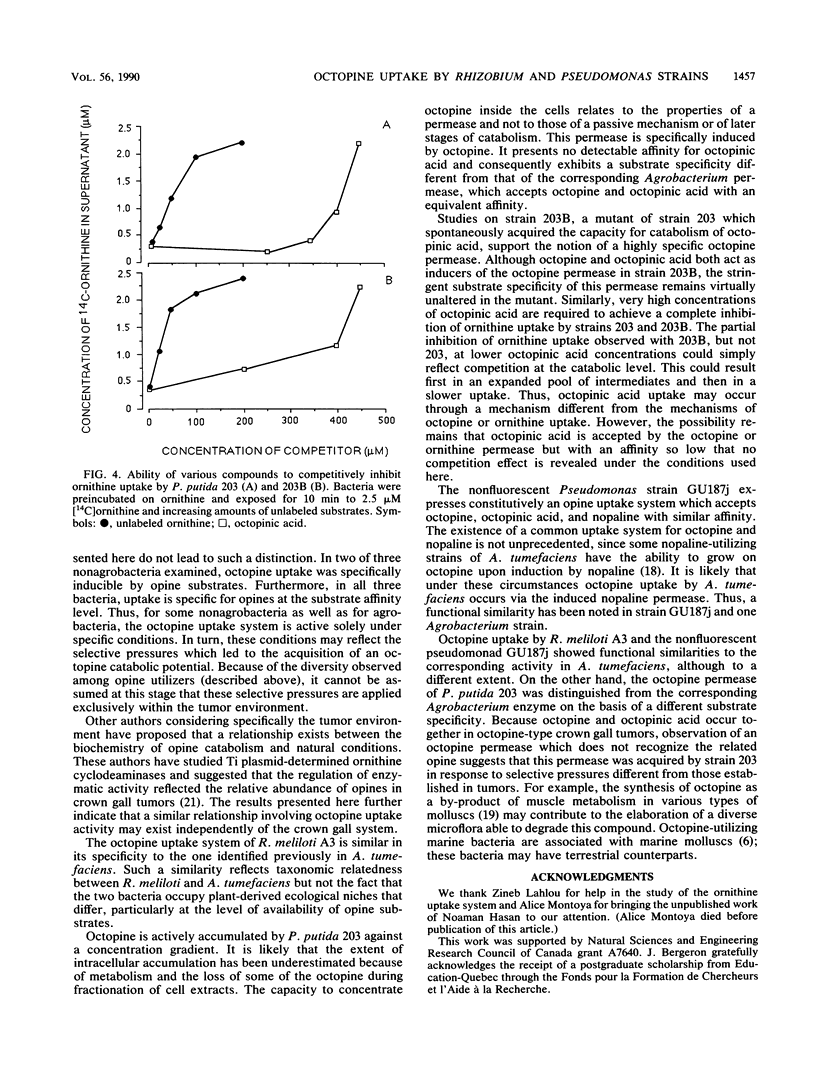
Abstract
The octopine-utilizing strain Agrobacterium tumefaciens B6S3 and three nonagrobacteria which had the capacity to utilize this opine were compared for octopine uptake. The characteristics of uptake by Rhizobium meliloti A3 and strain B6S3 were similar. In both bacteria, uptake activity was inducible by octopine and by the related opine octopinic acid, and competition assays showed that these two opine substrates were accepted by the same uptake system with an equivalent affinity. Cells of Pseudomonas putida 203 accumulated octopine against a concentration gradient, and this activity was induced specifically by octopine. While strain 203 did not utilize octopinic acid, a spontaneous mutant with a combined capacity for octopine and octopinic acid utilization was obtained. Both opines induced octopine uptake by this mutant, but octopinic acid was not a substrate for the induced system. Thus, the Pseudomonas uptake system exhibited a different specificity for octopine than the corresponding Agrobacterium system. The nonfluorescent pseudomonad GU187j, which utilized the three related opines octopine, octopinic acid, and nopaline, was constitutive for octopine uptake. Strain GU187j possessed a system which accepted these three opines, but not arginine or ornithine, with a similar affinity.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beauchamp C. J., Chilton W. S., Dion P., Antoun H. Fungal catabolism of crown gall opines. Appl Environ Microbiol. 1990 Jan;56(1):150–155. doi: 10.1128/aem.56.1.150-155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droniuk R., Wong P. T., Wisse G., Macleod R. A. Variation in Quantitative Requirements for Na for Transport of Metabolizable Compounds by the Marine Bacteria Alteromonas haloplanktis 214 and Vibrio fischeri. Appl Environ Microbiol. 1987 Jul;53(7):1487–1495. doi: 10.1128/aem.53.7.1487-1495.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon P., Chilton M. D., Petit A., Tempé J. Agropine in "null-type" crown gall tumors: Evidence for generality of the opine concept. Proc Natl Acad Sci U S A. 1980 May;77(5):2693–2697. doi: 10.1073/pnas.77.5.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapwijk P. M., Hooykaas P. J., Kester H. C., Schilperoort R. A., RORSCH A. Isolation and characterization of Agrobacterium tumefaciens mutants affected in the utilization of octopine, octopinic acid and lysopine. J Gen Microbiol. 1976 Sep;96(1):155–163. doi: 10.1099/00221287-96-1-155. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Murphy P. J., Heycke N., Banfalvi Z., Tate M. E., de Bruijn F., Kondorosi A., Tempé J., Schell J. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked and on the Sym plasmid. Proc Natl Acad Sci U S A. 1987 Jan;84(2):493–497. doi: 10.1073/pnas.84.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten L. A., Schilperoort R. A. A rapid micro scale method for the detection of lysopine and nopaline dehydrogenase activities. Biochim Biophys Acta. 1978 Dec 8;527(2):497–500. doi: 10.1016/0005-2744(78)90363-7. [DOI] [PubMed] [Google Scholar]
- ROBIN Y., NGUYEN VAN THOAI [Metabolism of guanidyl derivatives. X. Metabolism of octopine: its biological role]. Biochim Biophys Acta. 1961 Sep 16;52:233–240. doi: 10.1016/0006-3002(61)90672-2. [DOI] [PubMed] [Google Scholar]
- Schindler U., Sans N., Schröder J. Ornithine cyclodeaminase from octopine Ti plasmid Ach5: identification, DNA sequence, enzyme properties, and comparison with gene and enzyme from nopaline Ti plasmid C58. J Bacteriol. 1989 Feb;171(2):847–854. doi: 10.1128/jb.171.2.847-854.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. B., Wilson R., Shaw G. J., Petit A., Tempe J. Biosynthesis and degradation of nodule-specific Rhizobium loti compounds in Lotus nodules. J Bacteriol. 1987 Jan;169(1):278–282. doi: 10.1128/jb.169.1.278-282.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepfer D., Goldmann A., Pamboukdjian N., Maille M., Lepingle A., Chevalier D., Dénarié J., Rosenberg C. A plasmid of Rhizobium meliloti 41 encodes catabolism of two compounds from root exudate of Calystegium sepium. J Bacteriol. 1988 Mar;170(3):1153–1161. doi: 10.1128/jb.170.3.1153-1161.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay G., Gagliardo R., Chilton W. S., Dion P. Diversity among Opine-Utilizing Bacteria: Identification of Coryneform Isolates. Appl Environ Microbiol. 1987 Jul;53(7):1519–1524. doi: 10.1128/aem.53.7.1519-1524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K., Krishnan M., Gould J. H., Smith R. H., Gelvin S. B. Opines stimulate induction of the vir genes of the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1989 Jul;171(7):3696–3703. doi: 10.1128/jb.171.7.3696-3703.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Itano H. Phenanthrenequinone as an analytical reagent for arginine and other monosubstituted guanidines. Biochim Biophys Acta. 1966 Dec 28;130(2):538–540. doi: 10.1016/0304-4165(66)90256-x. [DOI] [PubMed] [Google Scholar]



