Abstract
Vitamin K-dependent (VKD) proteins require modification by the VKD-γ-glutamyl carboxylase, an enzyme that converts clusters of glus to glas in a reaction that requires vitamin K hydroquinone, for their activity. We have discovered that the carboxylase also carboxylates itself in a reaction dependent on vitamin K. When pure human recombinant carboxylase was incubated in vitro with 14CO2 and then analyzed after SDS/PAGE, a radiolabeled band corresponding to the size of the carboxylase was observed. Subsequent gla analysis of in vitro-modified carboxylase by base hydrolysis and HPLC showed that all of the radioactivity could be attributed to gla residues. Quantitation of gla, asp, and glu residues indicated 3 mol gla/mol carboxylase. Radiolabeled gla was acid-labile, confirming its identity, and was not observed if vitamin K was not included in the in vitro reaction. Carboxylase carboxylation also was detected in baculovirus(carboxylase)-infected insect cells but not in mock-infected insect cells, which do not express endogenous VKD proteins or carboxylase. Finally, we showed that the carboxylase was carboxylated in vivo. Carboxylase was purified from recombinant carboxylase BHK cells cultured in the presence or absence of vitamin K and analyzed for gla residues. Carboxylation of the carboxylase only was observed with carboxylase isolated from BHK cells cultured in vitamin K, and 3 mol gla/mol carboxylase were detected. Analyses of carboxylase and factor IX carboxylation in vitro suggest a possible role for carboxylase carboxylation in factor IX turnover, and in vivo studies suggest a potential role in carboxylase stability. The discovery of carboxylase carboxylation has broad implications for the mechanism of VKD protein carboxylation and Warfarin-based anti-coagulant therapies that need to be considered both retrospectively and in the future.
Vitamin K-dependent (VKD) proteins undergo an unusual posttranslational modification required for their biological activity, namely the carboxylation of clusters of glu residues to γ-glutamyl glus, or glas, in a region of the VKD proteins called the gla domain (1, 2). Carboxylation of the VKD proteins effects their Ca2+-mediated interaction with phospholipid bilayers and is carried out by an integral membrane endoplasmic reticulum enzyme, the VKD-γ-glutamyl carboxylase. The carboxylase modifies VKD substrates by using CO2, O2, and vitamin K hydroquinone as cofactors, and the carboxylase is also an epoxidase, converting the vitamin K hydroquinone to vitamin K 2,3-epoxide. Subsequent regeneration of the vitamin K hydroquinone cofactor is carried out by a reductase that has been characterized but not yet identified (3, 4). Carboxylation of the gla domain involves the modification of multiple glus, ranging from 3 to 12 for the different VKD proteins. This multiplicity raises the question of whether the carboxylase is a processive enzyme, i.e., effecting all modifications as a result of a single binding event. When purified bovine liver carboxylase was coincubated with an excess of a substrate derived from the factor IX (fIX) propeptide plus gla domain and the extent of fIX carboxylation was analyzed, some fully carboxylated fIX was observed among many intermediate forms, suggesting some degree of processivity (5).
The known VKD proteins presently comprise a family of ≈1 dozen proteins. Although the first VKD proteins identified were ones involved in hemostasis, VKD proteins involved in bone morphogenesis and in growth regulation also have been identified (6–8). Identification of the cDNA sequences for these VKD proteins revealed a homologous peptide, which is not observed in noncarboxylated proteins, that subsequently was shown by mutational analysis to be a recognition sequence for the carboxylase (9, 10). In most cases, this peptide is a propeptide that is cleaved from the VKD proteins during their secretion. There is also a limited amount of homology among the VKD proteins within the gla domain. Most of the known VKD proteins (factor VII, fIX, factor X, protein S, protein C, protein Z, prothrombin, and gas6) have very similar sequences in the gla domain whereas the matrix gla protein and bone gla protein are more divergent. However, even the bone gla protein and matrix gla protein share some homology with the remaining VKD proteins over an ≈7-aa stretch in the gla domain.
When assessed by using an assay measuring the carboxylation of small peptide analog substrates derived from the gla domain, γ-glutamyl carboxylase peptide activity has been detected in most tissues examined in higher organisms. The carboxylase has been purified from bovine liver and from the human 293 cell line (11–13), and a cDNA encoding the human carboxylase has been isolated (14). This cDNA encodes a 95-kDa protein confirmed to be the carboxylase by its ability to effect peptide activity when expressed in baculovirus-infected insect cells, which do not otherwise contain carboxylase activity (15). The cDNA sequence for the carboxylase does not predict any functional domains: A weak homology to lipoxygenases was detected, and some homology to the carboxylase recognition sequence of matrix gla protein (but not to any other VKD protein) was reported (14, 16). It was therefore surprising when we discovered that the carboxylase itself is a VKD protein. The evidence demonstrating carboxylase carboxylation is presented here, along with potential functional consequences of this newly identified carboxylase modification.
MATERIALS AND METHODS
Generation of r-Carboxylase Cell Lines.
A stably transfected cell line overexpressing carboxylase activity was generated by transfecting the plasmid r-carb/ZEM229, which contains a human r-carboxylase cDNA in the vector ZEM229 (17), into BHK cells. After selection in 1 μM methotrexate, individual colonies were screened for r-carboxylase expression by a carboxylase peptide activity assay (18) as well as by Western blot analysis using an α-carboxylase antibody (Ab) (see below).
A BHK cell line stably expressing r-fIX and overexpressing carboxylase activity by 70-fold was generated by cotransfecting the plasmids r-fIX/ZEM229, which contains the human r-fIX cDNA in the vector ZEM229, and r-carb/ZEM228, containing the human r-carboxylase cDNA in the vector ZEM228 (19). After selection in 0.5 mg/ml G418 and 150 nM methotrexate, colonies were screened by using an ELISA on secreted fIX (11) and a peptide carboxylase activity assay on cell lysates (18).
Purification of the Carboxylase.
Carboxylase was purified from r-carboxylase BHK cells cultured in the presence or absence of vitamin K (5 μg/ml) after preparing microsomes from 109 cells, as described (11). The microsomes (10 ml, 15 mg) were solubilized in 50 mM Tris (pH 7.4), 200 mM NaCl, 0.3% CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate}, and 2 mM phenylmethylsulfonyl fluoride by rocking for 1 h followed by centrifugation for 1 h at 45K rpm, all at 4°C. The supernatant was adsorbed overnight at 4°C to an α-C-terminal carboxylase peptide Ab coupled to CNBr-activated Sepharose (1 ml, 5 mg/ml of affinity-purified Ab per milliliter of resin; the C-terminal peptide comprised aa 744–758 of the human carboxylase). The resins were washed at room temperature as described (11), and the carboxylase was eluted overnight at room temperature by using a mixture of 100 μM C-terminal peptide and 100 μM factor X propeptide (added to stabilize the carboxylase) in 50 mM Tris (pH 7.4), 500 mM NaCl, 0.05% CHAPS, 0.05% phosphatidyl choline, and 5 mM DTT. All fractions were monitored for carboxylase by using a peptide assay (see below), and the peptide eluants were stored at −80°C.
To purify the carboxylase from r-carboxylase, r-fIX BHK cells, microsomes (30 ml, 60 mg) prepared from 4 × 109 cells cultured without vitamin K, were solubilized for 1 h in 50 mM Tris (pH 7.4), 200 mM NaCl, 0.5% CHAPS, and 2 mM phenylmethylsulfonyl fluoride, followed by centrifugation at 45,000 rpm for 1 h at 4°C. The supernatant was adsorbed to an α-fIX column, and carboxylase was eluted with propeptide as described (11). The propeptide eluant was assayed and stored at −80°C. To show that no fIX was present in the propeptide eluant, aliquots from several preparations were tested in an ELISA (11). Purified carboxylase preparations also were tested for fIX in Western blot analysis using affinity-purified polyclonal α-fIX (11) and purified plasma fIX (5–50 ng, Enzyme Research Laboratories, South Bend, IN) as a control.
Baculovirus containing the human r-carboxylase cDNA was generated as described (18). Insect cell lysates containing r-carboxylase were prepared by infecting SF-21 cells (2.5 × 107) with baculovirus(carboxylase) (multiplicity of infection = 10) and harvesting the cells 42 h postinfection (18). All steps were performed at 4°C. Cells were dislodged by scraping, rinsed twice in PBS (10 ml), and resuspended in 5 ml 25 mM imidazole (pH 7.3), 250 mM sucrose, and 2 mM phenylmethylsulfonyl fluoride. After sonication, the lysates were centrifuged at 3500 rpm for 15 min, and the supernatants were stored at −80°C.
Carboxylation Assays.
Peptide carboxylation was assayed in a 150-μl mixture of 0.06% CHAPS, 0.06% phosphatidyl choline, 0.06% sodium cholate, 1 M ammonium sulfate, 5 mM DTT, 2.5 mM NaH14CO3 (40–50 mCi/mmol), 50 mM BES {N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (pH 7.2)}, 5 mM Boc-Glu-Glu-Leu-Ome, 20 μM factor X propeptide, and 10 μg of vitamin K hydroquinone. A similar mixture lacking the Boc-Glu-Glu-Leu-Ome and propeptide was used for protein carboxylation. Carboxylation of the fIX–carboxylase complex was analyzed after isolating the complex from r-carboxylase, r-fIX BHK microsomes, which were found to contain ≈95% of the carboxylase in complex with the propeptide form of fIX (data not shown). Solubilized microsomes (1 ml, 2 mg) were adsorbed to α-C-terminal carboxylase peptide Ab resin (100 μl, 5 mg/ml) and then washed as described (11). An aliquot (2%) of the resin was assayed for peptide carboxylase activity to determine the amount of carboxylase by using a specific activity of 7 × 105 cpm/h/μg. The remaining resin was incubated in 400 μl of a protein carboxylation mix for varying times after the addition of vitamin K hydroquinone (20 μg), and the reaction was quenched by the addition of SDS/PAGE loading buffer. Samples were electrophoresed, and the gels were washed at least five times in 40% methanol/5% acetic acid, then dried and quantitated by PhosphorImager (Molecular Dynamics) analysis. 14C-methylated BSA standards were included on each gel for quantitation. Carboxylation of free fIX by pure carboxylase was analyzed similarly: A pure population of propeptide-containing, full length fIX (profIX) was isolated from a BHK cell line expressing high levels (15 μg/ml/day) of human r-fIX. As will be presented in detail elsewhere, the mixture of mature fIX and profIX secreted from this line was chromatographed over an α-fIX propeptide Ab column, and pure profIX was obtained, as assessed by N-terminal sequence analysis. ProfIX (850 nM) was preincubated with pure carboxylase (35 nM) for 2 h, and, at timed intervals after the addition of vitamin K hydroquinone (20 μg), samples were withdrawn and processed as above.
Gla Quantitation.
To monitor in vitro carboxylation, propeptide eluant from r-fIX, r-carboxylase BHK cells was chromatographed on Q-Sepharose, and the eluant (100 pmol) was incubated for 10 min in a protein carboxylation reaction (800 μl) with or without vitamin K (50 μg) followed by precipitation with 30% trichloroacetic acid (TCA). After centrifugation and two washes with 100% ethanol, the precipitates were resuspended in 100 μl 2.5 N KOH and hydrolyzed and chromatographed over Dowex columns (20). The samples were lyophilized twice, then resuspended in water and derivatized with o-phthalaldehyde (20). Samples were injected onto a C-18 column (ODS hypersil 5 μ, 100 × 2.1 mm, Hewlett–Packard), and a linear gradient of 5–50% acetonitrile in 10 mM sodium citrate (pH 7) was applied.
To monitor in vivo carboxylation, carboxylase purified from r-carboxylase BHK cells (20 pmol) cultured in the presence or absence of vitamin K was TCA precipitated (800 μl peptide eluant plus 400 μl TCA) and processed as above.
RESULTS
The Carboxylase Is Carboxylated in Vitro.
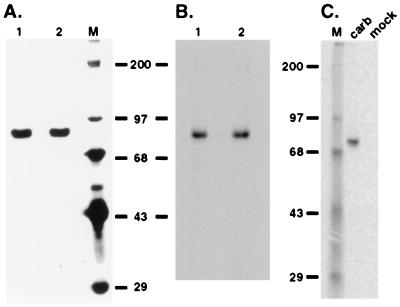
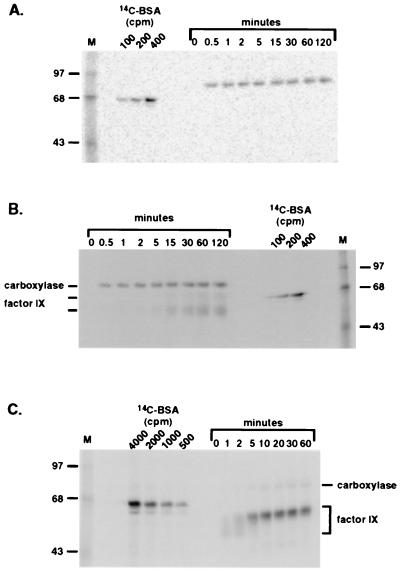
Pure carboxylase isolated from cells cultured in the absence of vitamin K incorporated 14CO2 when incubated in an in vitro protein carboxylation reaction (Fig. 1). Incorporation depended on vitamin K in the reaction (data not shown). The carboxylase preparations were homogenous, showing one band of the expected molecular mass (95 kDa, Fig. 1A), so incorporation of 14CO2 was apparently into the carboxylase and not some contaminant protein.
Figure 1.
In vitro modification of the carboxylase. Carboxylase was purified from a r-fIX, r-carboxylase BHK cell line. Two independent preparations (lanes 1 and 2, 4 × 106 cpm/h of peptide activity or ≈6 μg) were analyzed by SDS/PAGE and silver staining (A) and by incubation in a protein carboxylation reaction followed by SDS/PAGE and PhosphorImager analysis (B). (C) Solubilized microsomes (100 μg) from baculovirus(carboxylase)-infected or mock-infected SF-21 cells were incubated in a protein carboxylation reaction for 2 min and then analyzed by SDS/PAGE and PhosphorImager analysis. In parallel experiments performed in a reaction lacking vitamin K, no radiolabeled bands were detected even with incubations up to 2 h (data not shown).
The carboxylase had been isolated from a fIX–carboxylase complex derived from r-fIX, r-carboxylase BHK cells: The fIX-carboxylase complex was adsorbed to an α-fIX Ab column, and the carboxylase subsequently was eluted by using a propeptide to displace it from fIX. The concentration of carboxylase in the purified preparations was ≈1 μM. fIX was not detected in several different preparations using an ELISA sufficiently sensitive to detect 10−4 μM fIX. Moreover, the molecular mass of the fIX in the fIX–carboxylase complex from which the carboxylase was purified was 50 kDa (shown below) and not the 95 kDa molecular mass detected (Fig. 1B). Nonetheless, to unequivocally demonstrate that the 95-kDa radiolabeled band was not caused by comigration of a hyperglycosylated form of fIX with the carboxylase, Western blot analysis was performed. When the pure carboxylase preparations (3 pmol, Fig. 1A) were analyzed by using conditions in which as little as 0.03 pmol fIX could be detected, no fIX cross-reacting material was observed (data not shown). Thus, the combined ELISA and Western analysis results showed that the radiolabeled band (Fig. 1B) was not fIX and that it was the carboxylase that was being modified by 14CO2. This conclusion was verified independently by purifying the carboxylase from r-carboxylase BHK cells that did not express any other VKD protein by using a completely different method of purification (shown below).
Carboxylase modification also was observed when the carboxylase was expressed in insect cells, which lack endogenous carboxylase (15). When solubilized microsomes from baculovirus(carboxylase)-infected insect cells were incubated in an in vitro carboxylation reaction, a single radiolabeled band was observed (Fig. 1C). This band was not detected in mock (Fig. 1C) or wild-type, baculovirus-infected, insect cell-derived microsomes, and detection depended on the inclusion of vitamin K in the in vitro reaction (data not shown). The radiolabeled band was quantitatively immunocaptured by an α-C-terminal carboxylase antipeptide Ab but not by a control Ab (against fIX; data not shown).
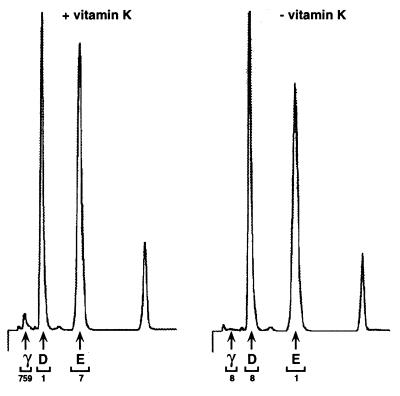
To determine whether the 14CO2 incorporation into the carboxylase was due to conversion of glus to glas, pure carboxylase (Fig. 1A) was incubated in an in vitro reaction, followed by base hydrolysis and subsequent resolution of amino acids by HPLC. As a control, a parallel sample was processed in which vitamin K was omitted from the in vitro reaction. As shown in Fig. 2, a peak migrating at the position expected for gla residues (0.8 min) was detected in a vitamin K-dependent manner. All of the radioactivity was recovered in the gla peak, with the cpm in this peak accounting for 98% of the cpm injected onto the C18 column. Quantitation of gla, asp, and glu peaks by fluorescence indicated an asp-to-glu ratio of 1.3, which would be predicted from the known sequence of the human carboxylase (14). The ratio of gla-to-asp indicated 3 mol of gla per mol of carboxylase. To confirm the identification of the peak at 0.8 min as a gla residue, in vitro carboxylated protein also was subjected to acid hydrolysis and HPLC analysis (data not shown). No peak at 0.8 min was detected, which was as expected because the gla residue is acid-labile (21). Thus, the 14C-covalent modification of the carboxylase observed in the in vitro carboxylation reactions (Fig. 1 B and C) was clearly due to carboxylation of the carboxylase.
Figure 2.
Gla analysis of in vitro carboxylated carboxylase. The propeptide eluant isolated from r-fIX, r-carboxylase BHK cells (100 pmol, Fig. 1) was chromatographed on Q-Sepharose, in vitro carboxylated, and then TCA precipitated and base hydrolyzed. Samples with or without vitamin K in the in vitro protein carboxylation reaction were processed in parallel. After the amino acid hydrolysate was resuspended in water (25 μl), duplicate aliquots (2 μl) were quantitated for radioactivity and duplicate samples (10 μl) were analyzed by HPLC. Peaks corresponding to gla (0.8 min), asp (1.6 min), and glu (3.5 min) were collected and counted, and the radioactivity (in cpm) is indicated at the bottom of the chromatogram.
Carboxylation of the Carboxylase in Vivo.
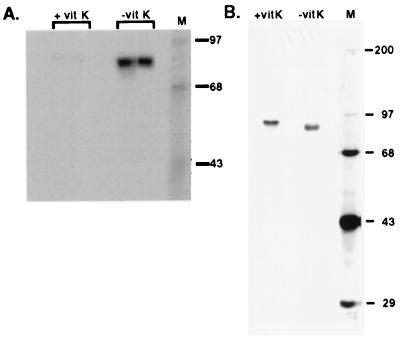
If the carboxylase is carboxylated in vivo, then carboxylase isolated from cells cultured in vitamin K-containing medium should already be carboxylated. This modification should preclude further incorporation of 14CO2 in vitro and be detectable by gla analysis. To determine whether the carboxylase was carboxylated in vivo, a stably transfected cell line overexpressing r-carboxylase by 50-fold was generated and analyzed. Both peptide activity and epoxidase activity also were overexpressed 50-fold, showing that the specific activity of the r-carboxylase was virtually identical to the endogenous carboxylase. Two sets of solubilized microsomes were prepared from this cell line, differing only in the addition of vitamin K to one set immediately before cell line scale up. When these solubilized microsomes were incubated in an in vitro carboxylation reaction and subsequently analyzed, a strong radiolabeled band was observed in microsomes from cells cultured without vitamin K whereas only a faint signal was observed with microsomes from cells cultured with vitamin K (Fig. 3A). The contrast in radiolabel detection was not caused by low levels of carboxylase in the microsomes derived from cells cultured in the presence of vitamin K: Western blot analysis of equivalent amounts of microsomal protein showed that the levels were not substantially different (data not shown). The difference in carboxylation in the two sets of microsomes strongly suggested that the carboxylase derived from cells cultured in vitamin K-containing medium was modified already.
Figure 3.
Analysis of carboxylase isolated from a r-carboxylase BHK cell line. (A) Solubilized microsomes were prepared from r-carboxylase BHK cells cultured in the presence or absence of vitamin K (vit K). Duplicate samples (50 μg) were incubated in an in vitro protein carboxylation reaction for 1 h and then processed by using SDS/PAGE and PhosphorImager analysis. (B) Carboxylase (≈200 ng) purified from the solubilized microsomes by chromatography on an α-C-terminal carboxylase peptide Ab column was analyzed by SDS/PAGE and silver staining.
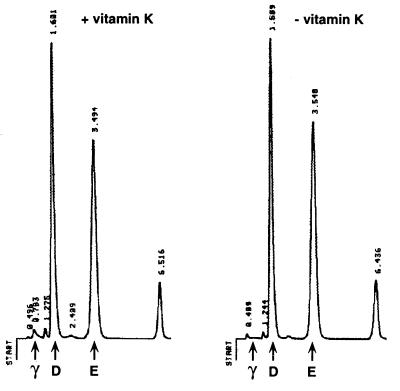
To directly assess carboxylase carboxylation in vivo, carboxylase was purified and subjected to gla analysis. Purification comprised the adsorption of solubilized microsomes to an α-C-terminal carboxylase peptide Ab column and subsequent elution with peptide. The peptide eluants were shown to be homogenous by SDS/PAGE and silver staining (Fig. 3B). The carboxylase isolated from cells cultured in vitamin K was ≈4 kDa larger than that from cells depleted in vitamin K, which could be due to the gla modifications or to some other posttranslational modification. When the peptide eluants were TCA precipitated, which removed free peptide, and subjected to gla quantitation, a peak corresponding to the position expected for gla was detected with the carboxylase purified from cells cultured in vitamin K (Fig. 4). In contrast, no gla peak was observed with carboxylase purified from cells cultured in the absence of vitamin K (Fig. 4). Quantitation of individual gla, asp, and glu peaks gave a ratio of 3 mol of gla per mol of carboxylase, and the ratio of asp-to-glu was as expected for the carboxylase. These results show that the carboxylase is carboxylated in vivo in a vitamin K-dependent manner and unequivocally demonstrate that carboxylase carboxylation was not an artifact of the in vitro reaction conditions.
Figure 4.
Gla analysis of carboxylase purified from r-carboxylase BHK cells. Carboxylase purified from r-carboxylase BHK cells cultured with or without vitamin K (20 pmol each, Fig. 3B) was TCA precipitated and base hydrolyzed. The amino acid hydrolysates were resuspended in 100 μl of water, and triplicate aliquots were analyzed by HPLC. The numbers above each peak indicate the retention times.
Kinetics of Carboxylase and fIX Carboxylation.
The rates of carboxylase or fIX carboxylation were compared in vitro for free carboxylase vs. free carboxylase plus profIX vs. a preformed carboxylase–fIX complex. The kinetics of carboxylation of free carboxylase were rapid, with the reaction plateauing within 1 min of adding vitamin K (Fig. 5A). The rate of carboxylase carboxylation when free carboxylase was incubated with profIX was virtually identical to that of free carboxylase alone; again, complete modification occurred within 1 min of the addition of vitamin K (Fig. 5B). Carboxylase carboxylation preceded fIX carboxylation; fIX carboxylation was not detected until ≈5 min (Fig. 5B). In contrast, when carboxylase carboxylation was analyzed by using a preformed fIX–carboxylase complex (Fig. 5C), carboxylase carboxylation was delayed and observed after fIX carboxylation. Carboxylase carboxylation was not detected until 5 min, corresponding to the time when fIX carboxylation plateaued. Thus, occupation of the carboxylase active site by fIX in the complex appeared to block carboxylase carboxylation.
Figure 5.
Kinetics of carboxylation of factor IX and carboxylase. (A and B) Pure carboxylase (Fig. 1, 14 pmol, equivalent to 106 cpm/h of peptide activity) was incubated (A) alone or (B) with profIX (340 pmol). (C) Carboxylase–fIX complex (equivalent to 2 × 106 cpm/h of peptide activity or 28 pmol carboxylase and presumably an equimolar amount of fIX) was isolated by using α-C-terminal carboxylase peptide Ab resin, as described in Materials and Methods. The fIX in the complex is the profIX form of fIX. All three assays were performed in a final volume of 400 μl of protein carboxylation mix, and at timed intervals after the addition of vitamin K hydroquinone (20 μg), aliquots (40 μl) were withdrawn and the reaction was quenched by the addition of 40 μl of SDS/PAGE loading buffer. The samples were then analyzed by SDS/PAGE and PhosphorImager analysis by using 14C-methylated BSA standards to convert the PhosphorImager signal to cpm. The 50- and 60-kDa profIX forms in B are indicated by two lines on the left.
The turnover of profIX gla residues when incubated with free carboxylase was ≈0.004 gla/s vs. ≈0.04–0.08 gla/s for profIX in the complex. These values were determined by using a peptide activity assay to quantitate the amount of carboxylase. Carboxylase in complex may have an ≈2-fold higher specific activity than free carboxylase (11), so at present the precise difference in rates (i.e., 10-fold or 20-fold for complex vs. free carboxylase plus profIX) is not known and will require a better assay for quantitating carboxylase.
The gel pattern of fIX carboxylation was also different when a preformed complex vs. free carboxylase plus profIX was analyzed. A ladder of fIX forms was observed at early times in the carboxylation of the complex (1 min, Fig. 5C), with the most abundant form having a molecular mass of ≈50 kDa. By 5 min, the most abundant form had shifted to a molecular mass of ≈60 kDa. This 60-kDa form is presumably the extensively, or fully, carboxylated fIX that migrates slower because of the increase in the number of negative charges (12 mol/mol fIX) altering SDS binding. In contrast, the incubation of free carboxylase with profIX resulted in only limited conversion of the 50-kDa fIX form to 60 kDa, even after much longer times of incubation (Fig. 5B). For example, when the 50-kDa band at 30 min and 60-kDa band at 60 min were quantitated, only 15% conversion was observed. However, fIX carboxylation clearly was occurring over the 2 h assayed, as evidenced by the linear increase in cpm into the 50-kDa fIX form over this period. This contrast in the efficient shift from 50 kDa to 60 kDa in the complex vs. limited conversion when free carboxylase was incubated with profIX suggests differences in processivity in the two assays.
DISCUSSION
We report the discovery of vitamin K-dependent carboxylase carboxylation. In vivo carboxylation was demonstrated by using gla analysis on pure carboxylase from r-carboxylase BHK cells (Figs. 3B and 4). In vitro carboxylase carboxylation also was demonstrated, both by gla quantitation on pure carboxylase (Fig. 2) and also by analyzing carboxylation in baculovirus-infected insect cells (Fig. 1C), which lack endogenous carboxylase. A total of 3 mol of gla per mol of carboxylase was indicated by both the in vitro and in vivo analyses (Figs. 2 and 4). Precedence for enzymatic self-modification exists in other systems, for example autophosphorylation of several protein kinases (22). However, a mechanism of self-carboxylation never has been proposed for the carboxylase and comes as a surprise, given the lack of homology between the carboxylase and the previously known VKD proteins. These family members all share two homologous regions, one comprising the recognition sequence for the carboxylase (23) and the other a portion of the gla domain. A limited amount of homology was observed between the carboxylase (aa 495–518 for the human homolog) and the carboxylase recognition sequence of the matrix gla protein but not of any of the other VKD proteins. In addition, the carboxylase does not share any apparent homology with the VKD proteins in their gla domain, and so there was no reason to predict carboxylase carboxylation.
Carboxylation of the carboxylase occurred in the absence of propeptide (Figs. 1C and 3A), which is of interest with respect to the demonstrated ability of the propeptide to activate peptide carboxylation (24) as well as to a proposed role for the propeptide in regulating protein carboxylation (16). Our results agree with previous ones showing that exogenously added propeptide can activate peptide activity (24): Peptide carboxylation in r-carboxylase BHK cells or in baculovirus(carboxylase)-infected insect cells was stimulated 40- or 100-fold, respectively, by the addition of propeptide to the in vitro reaction (data not shown). However, carboxylase carboxylation was observed in microsomes from both sources, despite the absence of propeptide (Figs. 1C and 3A).
As described above, a region within the carboxylase with potential homology to the matrix gla protein carboxylase recognition sequence was identified (16), and a model was proposed in which this part of the carboxylase acts as an inhibitory domain, shielding the active site and preventing promiscuous carboxylation of non-VKD proteins. The VKD proteins, which contain the carboxylase recognition sequence, would compete with this carboxylase sequence, thereby allowing their modification by the carboxylase. This model of the carboxylase existing in a closed conformation in the absence of propeptide has not yet been tested; however, our results clearly show that carboxylase carboxylation does not require the putative open conformation. Carboxylase carboxylation in a closed conformation could be consistent with the model if it occurs by an intramolecular mechanism, which is very likely the case. We found that the rate of carboxylase carboxylation was independent of the enzyme concentration (data not shown), suggesting cis-carboxylation of the carboxylase.
The difference in kinetics of fIX and carboxylase carboxylation observed when the two were in a complex vs. added as separate components may provide insight into the function of carboxylase carboxylation, i.e., in regulating the carboxylation of the VKD proteins. When carboxylase was assayed either alone or when coincubated with profIX, it was carboxylated within the first minute of the reaction (Fig. 5 A and B). In contrast, when analyzed by using a preformed carboxylase–fIX complex, carboxylase carboxylation was delayed, and the onset of carboxylase carboxylation correlated with the apparent completion of fIX carboxylation (Fig. 5C). Thus, fIX bound to the carboxylase active site blocked carboxylase carboxylation. After fIX carboxylation, the active site is freed, allowing subsequent access of glu residues in the carboxylase for modification. Carboxylation of the carboxylase may facilitate the release of the fIX product: The introduction of three negative charges into the carboxylase could result in repulsion between the carboxylase and the anionic fIX gla domain.
An increase in fIX release due to carboxylase carboxylation could manifest in decreased processivity of the carboxylase and could account for the differences in fIX carboxylation we observed when fIX was analyzed as an independent component vs. a preformed complex with carboxylase (Fig. 5 B and C). When profIX and carboxylase were incubated together, carboxylase carboxylation was shown to precede fIX carboxylation (Fig. 5B). Thus, the fIX was being modified by a carboxylated carboxylase in this assay. In contrast, when fIX was analyzed in a fIX–carboxylase complex, the fIX was shown to be modified by an uncarboxylated carboxylase (Fig. 5C). Two significant differences were observed in the carboxylation of fIX when analyzed as an independent component vs. in a preformed complex. First, the rate of profIX carboxylation when incubated with free carboxylase (Fig. 5B) was extremely slow, with a turnover (0.004 gla/s) 2–3 logs lower than observed for the peptide substrate Boc-Glu-Glu-Leu-Ome (≈1 gla/s) (11) and a rate 10- to 20-fold lower than for carboxylation in the complex (Fig. 5C). Second, the gel pattern of fIX carboxylation was different in the two assays. FIX in the complex showed a dramatic and complete shift in molecular mass forms during the reaction (Fig. 5C), suggesting a highly processive mechanism of fIX carboxylation that converts fIX to the near or fully carboxylated form. However, when free profIX was incubated with carboxylase (Fig. 5B), only a limited amount of the fIX was shifted to the higher molecular mass form. This limited conversion is consistent with the results obtained by Morris et al. (5); when bovine liver carboxylase was incubated with a fIX derivative substrate and the products were analyzed, most of the fIX product was undercarboxylated (i.e., 1–11 glas per mol fIX), and only a small amount of fully carboxylated fIX was observed (5). The contrast in fIX carboxylation that we observed in the two assays is consistent with differences in processivity, and these differences correlate with the carboxylation of the carboxylase. Thus, the carboxylated carboxylase may increase the rate of release of partially carboxylated fIX, resulting in decreased processivity and accounting for the differences in fIX carboxylation observed (Fig. 5 B vs. C). We currently are testing the processivity of carboxylase on fIX when the two are in a complex or added as separate components as a first step toward assessing the potential role of carboxylase carboxylation in product release and/or processivity.
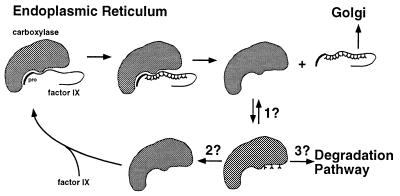
The discovery of carboxylase carboxylation has potentially broad implications with respect to the mechanism of VKD protein carboxylation, which need to be considered in future experiments as well as retrospectively. All previous in vitro carboxylation studies have been performed by using a count-based assay, and so the state of carboxylase carboxylation cannot be reevaluated. The rapid rate of carboxylation that we observed with pure carboxylase (Fig. 5) as well as in crude systems (Fig. 1C), however, would argue that, regardless of the source of carboxylase (i.e., whether from Warfarin-treated or vitamin K-containing or lacking cells or tissue), with all previous in vitro carboxylation assays, the carboxylase was most likely carboxylated at the beginning of the reaction. The identification of carboxylase carboxylation now introduces a layer of complexity that invokes a substantial number of new questions (Fig. 6). In addition to the questions already addressed above, i.e., whether carboxylase carboxylation affects release and/or processivity of VKD proteins, there is also the question of the steady–state levels of carboxylated carboxylase in vivo; in contrast to in vitro conditions, in which the availability of vitamin K can be controlled, under normal physiological in vivo conditions, there is always a constant supply of vitamin K. There are also the issues of whether carboxylase carboxylation occurs by an intramolecular mechanism and whether it occurs during each turnover of a VKD protein. If carboxylase carboxylation occurs during each turnover, then there is the question of whether decarboxylation is required for a subsequent enzymatic cycle. If required, then the mechanism for decarboxylation becomes an issue: Is decarboxylation due to reversibility of the reaction or mediated by another enzyme? How is decarboxylation specific only for the carboxylase? Finally, we recently have found that carboxylase carboxylation decreases the half-life of the carboxylase in vivo (data not shown), suggesting an additional role for this modification in carboxylase stability. This result was obtained by using the r-carboxylase BHK cell line that does not express any other VKD proteins, raising the questions of whether the presence of other VKD proteins affects the extent of carboxylase carboxylation and consequent carboxylase stability in vivo and what mechanism accounts for an effect of carboxylation on carboxylase stability. Combined in vivo analyses of the carboxylase as well as mutational analysis to block carboxylase carboxylation should provide powerful tools for addressing these new questions.
Figure 6.
New questions raised by the discovery of carboxylase carboxylation. The carboxylation of fIX and the carboxylase are depicted. The enzymatic properties and fate of the carboxylated carboxylase are open questions, which include whether carboxylase carboxylation affects the stability of the carboxylase (3?) and whether carboxylase carboxylation and decarboxylation occur during the turnover of each VKD substrate (2?). The existence of a decarboxylase activity, either by reaction reversibility (1?) or by an unidentified decarboxylase (2?), is also unknown.
Acknowledgments
We thank Beth McNally and Susan Lingenfelter for generating the r-carboxylase and r-carboxylase, r-fIX cell lines and Sadu Karnik and Kurt Runge for their helpful comments in the preparation of this manuscript. This work was supported by National Institutes of Health Grant HL55666 to K.L.B.
ABBREVIATIONS
- VKD
vitamin K-dependent
- r-carboxylase
recombinant carboxylase
- Ab
antibody
- fIX
factor IX
- r-fIX
recombinant factor IX
- profIX
propeptide-containing, full length fIX
- TCA
trichloroacetic acid
References
- 1.Suttie J W. Annu Rev Biochem. 1985;54:459–477. doi: 10.1146/annurev.bi.54.070185.002331. [DOI] [PubMed] [Google Scholar]
- 2.Furie B, Furie B C. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 3.Wallin R, Martin L F. J Clin Invest. 1985;76:1879–1884. doi: 10.1172/JCI112182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell R G, Matschiner J T. Arch Biochem Biophys. 1970;141:473–476. doi: 10.1016/0003-9861(70)90164-5. [DOI] [PubMed] [Google Scholar]
- 5.Morris D P, Stevens R D, Wright D J, Stafford D W. J Biol Chem. 1995;270:30491–30498. doi: 10.1074/jbc.270.51.30491. [DOI] [PubMed] [Google Scholar]
- 6.Varnum B C, Young C, Elliott G, Garcia A, Bartley T D, Fridell Y W, Hunt R W, Trail G, Clogston C, Toso R U, Yanagihara D, Bennett L, Sylber M, Merewether L A, Tseng A, Escobar E, Liu E T, Yamane H K. Nature (London) 1995;373:623–626. doi: 10.1038/373623a0. [DOI] [PubMed] [Google Scholar]
- 7.Manfioletti G, Brancolini C, Avanzi G, Schneider C. Mol Cell Biol. 1993;13:4976–4985. doi: 10.1128/mcb.13.8.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price P A. Annu Rev Nutr. 1988;8:565–583. doi: 10.1146/annurev.nu.08.070188.003025. [DOI] [PubMed] [Google Scholar]
- 9.Foster D C, Rudinski M S, Schach B G, Berkner K L, Kumar A A, Hagen F S, Sprecher C A, Insley M Y, Davie E W. Biochemistry. 1987;26:7003–7011. doi: 10.1021/bi00396a022. [DOI] [PubMed] [Google Scholar]
- 10.Jorgensen M J, Cantor A B, Furie B C, Brown C L, Shoemaker C B, Furie B. Cell. 1987;48:185–191. doi: 10.1016/0092-8674(87)90422-3. [DOI] [PubMed] [Google Scholar]
- 11.Lingenfelter S E, Berkner K L. Biochemistry. 1996;35:8234–8243. doi: 10.1021/bi9523318. [DOI] [PubMed] [Google Scholar]
- 12.Berkner K L, Harbeck M, Lingenfelter S, Bailey C, Sanders-Hinck C M, Suttie J W. Proc Natl Acad Sci USA. 1992;89:6242–6246. doi: 10.1073/pnas.89.14.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S M, Morris D P, Stafford D W. Proc Natl Acad Sci USA. 1991;88:2236–2240. doi: 10.1073/pnas.88.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S M, Cheung W F, Frazier D, Stafford D W. Science. 1991;254:1634–1636. doi: 10.1126/science.1749935. [DOI] [PubMed] [Google Scholar]
- 15.Roth D A, Rehemtulla A, Kaufman R J, Walsh C T, Furie B, Furie B C. Proc Natl Acad Sci USA. 1993;90:8372–8376. doi: 10.1073/pnas.90.18.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price P A, Williamson M K. Protein Sci. 1993;2:1987–1988. doi: 10.1002/pro.5560021120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster D C, Holly R D, Sprecher C A, Walker K M, Kumar A A. Biochemistry. 1991;30:367–372. doi: 10.1021/bi00216a009. [DOI] [PubMed] [Google Scholar]
- 18.Berkner K L, McNally B A. Methods Enzymol. 1997;282:313–333. doi: 10.1016/s0076-6879(97)82117-9. [DOI] [PubMed] [Google Scholar]
- 19.Busby S J, Mulvihill E, Rao D, Kumar A A, Lioubin P, Heipel M, Sprecher C, Halfpap L, Prunkard D, Gambee J, Foster D C. J Biol Chem. 1991;266:15286–15292. [PubMed] [Google Scholar]
- 20.Berkner K L. Methods Enzymol. 1993;222:450–477. doi: 10.1016/0076-6879(93)22029-f. [DOI] [PubMed] [Google Scholar]
- 21.Hauschka P V. Anal Biochem. 1977;80:212–223. doi: 10.1016/0003-2697(77)90640-6. [DOI] [PubMed] [Google Scholar]
- 22.van der Geer P, Hunter T, Lindberg R A. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 23.Pan L C, Price P A. Proc Natl Acad Sci USA. 1985;82:6109–6113. doi: 10.1073/pnas.82.18.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knobloch J E, Suttie J W. J Biol Chem. 1987;262:15334–15337. [PubMed] [Google Scholar]