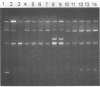
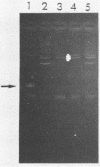
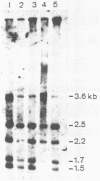
Abstract
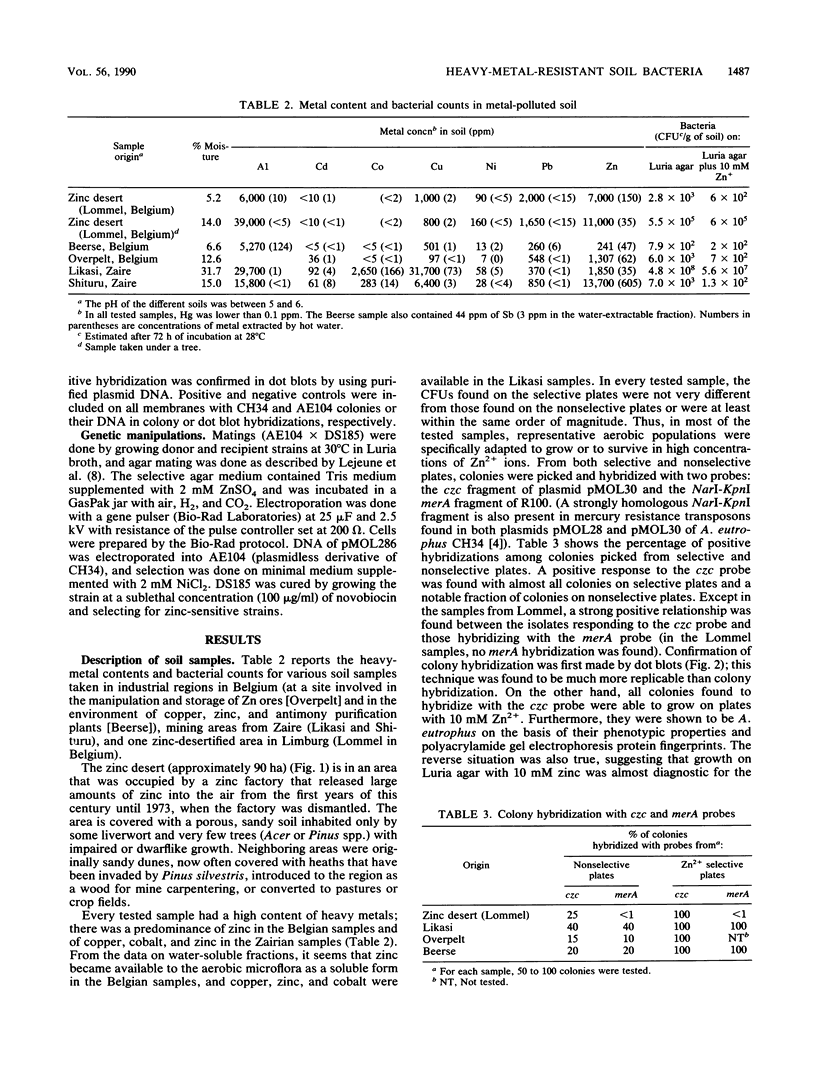
Alcaligenes eutrophus CH34 DNA fragments encoding resistance to Cd2+, Co2+, Zn2+ (czc), or Hg2+ (merA) were cloned and used as probes in colony hybridization procedures with bacteria isolated from polluted environments such as a zinc factory area (desertified because of the toxic effects of zinc contamination) and from sediments from factories of nonferrous metallurgy in Belgium and mine areas in Zaire. From the different soil samples, strains could be isolated and hybridized with the czc probe (resistance to Cd2+, Co2+, and Zn2+ from plasmid pMOL30). Percentages of CFU isolated on nonselective plates which hybridized with the czc and the mercury resistance probes were, respectively, 25 and 0% for the zinc desert, 15 to 20 and 10 to 20% for the two Belgian factories, and 40 and 40% for the Likasi mine area. Most of these strains also carried two large plasmids of about the same size as those of A. eutrophus CH34 and shared many phenotypic traits with this strain. These findings indicated a certain correlation between the heavy-metal content in contaminated soils and the presence of heavy-metal-resistant megaplasmid-bearing A. eutrophus strains.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy P. S., Hiatt H. D. Survival and detection of bacteria in an aquatic environment. Appl Environ Microbiol. 1989 Apr;55(4):788–793. doi: 10.1128/aem.55.4.788-793.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Fouts D. L., Olson B. H. Preparation of a DNA gene probe for detection of mercury resistance genes in gram-negative bacterial communities. Appl Environ Microbiol. 1985 Mar;49(3):686–692. doi: 10.1128/aem.49.3.686-692.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Tripp S. C., Olson B. H. Effect of metal-rich sewage sludge application on the bacterial communities of grasslands. Appl Environ Microbiol. 1985 Feb;49(2):333–337. doi: 10.1128/aem.49.2.333-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune P., Mergeay M., Van Gijsegem F., Faelen M., Gerits J., Toussaint A. Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4::mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol. 1983 Sep;155(3):1015–1026. doi: 10.1128/jb.155.3.1015-1026.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Houba C., Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas [proceedings]. Arch Int Physiol Biochim. 1978 May;86(2):440–442. [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H., Nies A., Chu L., Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H., Silver S. Metal ion uptake by a plasmid-free metal-sensitive Alcaligenes eutrophus strain. J Bacteriol. 1989 Jul;171(7):4073–4075. doi: 10.1128/jb.171.7.4073-4075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H., Silver S. Plasmid-determined inducible efflux is responsible for resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989 Feb;171(2):896–900. doi: 10.1128/jb.171.2.896-900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D., Mergeay M., Friedrich B., Schlegel H. G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987 Oct;169(10):4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHATZ A., BOVELL C., Jr Growth and hydrogenase activity of a new bacterium, Hydrogenomonas facilis. J Bacteriol. 1952 Jan;63(1):87–98. doi: 10.1128/jb.63.1.87-98.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Shields M. S., Tedford E. T., Breen A., Hooper S. W., Sirotkin K. M., Davis J. W. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol. 1985 May;49(5):1295–1303. doi: 10.1128/aem.49.5.1295-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Zelibor J. L., Jr, Doughten M. W., Grimes D. J., Colwell R. R. Testing for bacterial resistance to arsenic in monitoring well water by the direct viable counting method. Appl Environ Microbiol. 1987 Dec;53(12):2929–2934. doi: 10.1128/aem.53.12.2929-2934.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]