Abstract
A cDNA encoding a β-1,4-galactosyltransferase named β-1,4-GalT II was cloned from a cDNA library of the human breast tumor cell line, MRK-nu-1. Initially, a 860-bp PCR fragment was obtained from MRK-nu-1 mRNA by 3′-rapid amplification of cDNA ends by using two nested degenerate oligonucleotide primers based on a highly conserved amino acid sequence found in the catalytic domain of mammalian β-1,4-galactosyltransferases and Lymnaea stagnalis β-1,4-N-acetylglucosaminyltransferase (β-1,4-GlcNAcT), both of which utilize the same sugar acceptor. This subsequently was used as a probe to isolate a 4.7-kb cDNA that contained an ORF of 1,164 bp predicting a polypeptide of 388 aa. Its deduced amino acid sequence shows an identity of 37% with that of the previously characterized human β-1,4-galactosyltransferase (referred to as β-1,4-GalT I) and of 28% with that of L. stagnalis β-1,4-GlcNAcT. Study of the properties of the β-1,4-GalT II fused to protein A expressed as a soluble form in COS-7 cells revealed that it is a genuine β-1,4-GalT but has no lactose synthetase activity in the presence of α-lactalbumin. Northern blot analysis of 24 human tissues showed that they all express the β-1,4-GalT II transcript, although the levels varied. These results indicate that human cells contain another β-1,4-GalT.
Keywords: 3′-rapid amplification of cDNA ends, PCR, glycosyltransferase, tissue distribution
Cell surface carbohydrates have been demonstrated to play important roles in many biological events. In particular, complex-type sugar chains on mammalian glycoconjugates containing Galβ1 → 4GlcNAc outer branches carrying polysialic acid and HNK-1 epitopes or LewisX and sialyl LewisX determinants seem to be involved in adhesion processes such as cell-to-cell and cell-to-matrix interactions (reviewed in refs. 1–4). The two enzymes controlling the biosynthesis of complex-type oligosaccharides, UDP-GlcNAc:Man β-1,2-N-acetylglucosaminyltransferase (GlcNAcT-I) and UDP-Gal:GlcNAc β-1,4-galactosyltransferase (β-1,4-GalT) [EC 2.4.1.38], are among the best-studied glycosyltransferases (5–9). Targeted inactivation of the mouse GlcNAcT-I gene has been shown to result in death of the mice at mid-embryonic stages, whereas ablation of the β-1,4-GalT gene resulted in growth retardation and early death of the mice, indicating that the absence of complex-type N-linked oligosaccharides carrying the carbohydrate antigens described above and/or terminal N-acetylglucosamine or galactose residues themselves have important functions in embryonic development and further growth of the individual animals (10–13). The β-1,4-GalT-deficient mice, however, were able to synthesize a reduced amount of complex-type oligosaccharides containing galactose and also show low residual β-1,4-GalT activity. It is possible that such mice survive the gestation because of a partial compensation of the defective glycosylation by an additional, so far unidentified, β-1,4-GalT. Most β-1,4-GalTs characterized so far show lactose synthetase activity in the presence of α-lactalbumin (14, 15). No lactose was found in the milk of the mutant mice (13), indicating that the predicted additional β-1,4-GalT may not have this property. Interestingly, the occurrence of β-1,4-GalTs that appear to be unresponsive to α-lactalbumin has been described in porcine trachea and in Schistosoma mansoni (16, 17).
Mammalian β-1,4-GalTs are structurally related to a Lymnaea stagnalis UDP-GlcNAc:GlcNAc β-1,4-N-acetylglucosaminyltransferase (β-1,4-GlcNAcT) (18), an enzyme that shows an acceptor–substrate requirement resembling that of β-1,4-GalT (19). By comparing the amino acid sequences deduced from mammalian β-1,4-GalT cDNAs (8, 20, 21), two putative chicken GalT cDNAs (22), L. stagnalis β-1,4-GlcNAcT cDNA (18), and two related Caenorhabditis elegans genes (23), several conserved amino acid sequences have been discovered in the putative catalytic domain (24). To explore whether additional β-1,4-GalTs occur in mammalian cells, a 3′-rapid amplification of cDNA ends (RACE) approach was used with degenerate PCR primers based on one of the conserved amino acid boxes (Y/F)(W/F)GWG(G/A)EDD(D/E). In the present study we describe the isolation of a human cDNA encoding a β-1,4-GalT, the properties and tissue distribution of which differ from those of the well known mammalian β-1,4-GalT.
MATERIALS AND METHODS
Materials, Cells, and Oligonucleotides.
UDP-Gal (48.8 Ci/mmol; 1 Ci = 37 GBq), UDP-GalNAc (24 Ci/mmol), and UDP-GlcNAc (18.9 Ci/mmol) were purchased from DuPont/NEN and diluted with unlabeled UDP-sugars (Sigma) to give the desired specific activity. GlcNAcβ1 → 2Manα1 → 3Manβ1 → O(CH2)8COOCH2-m-nitrophenol (mNP), GlcNAcβ1 → 4(GlcNAcβ1 → 2)Manα1 → 3Manβ1 → O(CH2)8COOCH2-mNP, and GlcNAcβ1 → 6(GlcNAcβ1 → 2)Manα1 → 3Manβ1 → O(CH2)8COOCH2-mNP, which were chemically synthesized and donated by T. Kitajima, Towa Kasei Co. (Japan), and p-nitrophenyl-N-acetyl-1-thio-β-d-glucosaminide (GlcNAcβ-S-pNP) was purchased from Sigma. Asialo (As), agalacto (Ag)-human transferrin was prepared as described previously (25). The human breast tumor cell line, MRK-nu-1, was established by M. Sekiguchi at the Institute of Medical Science, University of Tokyo (Japan). The oligonucleotides, TS11: 5′-GTCCCGGAATTCCGGGCAAGGCATTCTGATCCGGGACAACGTGAGA-3′, TS9: 5′-CCGGAATTCCGGCTCCTCTCAGTACTCGTTCACCTGAGCCAGCTC-3′, HB25: 5′-TAIT(G/T)(C/G/T)GGITGGGG-3′, and ID10: 5′-GGITGGGGIIIIGA(A/G)GA(C/T)GA(C/T)GA-3′, were synthesized by Nippon Flour Mills (Kanagawa, Japan).
Isolation of a β-1,4-GalT-Related PCR Fragment by 3′-RACE.
Poly(A)+ RNA was prepared from MRK-nu-1 cells with a Fast Track mRNA Isolation kit (Invitrogen) and used for first-strand cDNA synthesis. Subsequently, this cDNA was used as a template with two nested primers, HB25 and ID10, in an adapted 3′-RACE protocol (GIBCO/BRL). A first-round of PCR [95°C, 3 min (94°C, 1 min; 50°C, 1 min; and 72°C, 1 min) × 25, and 72°C, 15 min] was carried out by using a tailed oligo(dT) primer already present in the cDNA sample mixture (25 nM) and primer HB25 (400 nM). In the second round of PCR, an aliquot of this reaction was subjected to PCR [95°C, 3 min (94°C, 1 min; 63°C, 1 min; and 72°C, 1 min) × 30, and 72°C, 15 min] by using primer ID10 (400 nM) and universal amplification primer (80 nM). Subsequently, the PCR fragments were separated by agarose gel electrophoresis, isolated from the gel, and purified with a Prep-A-Gene DNA purification kit (Bio-Rad), cloned into pBluescript SK (Stratagene), and sequenced.
Isolation of a Human cDNA Clone.
A cDNA library was constructed from the poly(A)+ RNA by using the λ-ZAP Express Vector Cloning kit (Stratagene). Approximately 3 × 105 independent clones were screened by using the 860-bp PCR fragment labeled with [α-32P]dCTP and by using a Ready to Go DNA labeling kit (Pharmacia) as a probe and hybridization conditions (60°C) as described previously (18). The nucleotide sequence of the cDNA was determined by the deoxynucleotide chain termination method (26) with an Auto Read Sequencing kit (Pharmacia).
Northern Blot Analysis of Human Tissues.
Human multiple tissue Northern blots, obtained from CLONTECH, were hybridized with part (nucleotides 145-1164) of the [α-32P]dCTP-labeled β-1,4-GalT II cDNA. Blots were subsequently reprobed with part (nucleotides 594-1200) of human β-1,4-GalT I cDNA, which was kindly supplied by M. N. Fukuda (The Burnham Institute, La Jolla, CA) and β-actin cDNA under the conditions as recommended by the manufacturer.
Construction of a Protein A/β-1,4-GalT II cDNA Fusion Plasmid.
A plasmid containing a fusion between part of the protein A sequence (27) and the β-1,4-GalT II cDNA was constructed as follows. A cDNA fragment in which the putative N-terminal cytoplasmic and transmembrane sequences had been removed was obtained by PCR by using the isolated cDNA as a template and the oligonucleotide primers TS11 and TS9. The resultant PCR fragment was sequenced, digested with EcoRI, and cloned into the EcoRI site of pPROTA (28), which resulted in the hybrid construct.
Expression of the Fusion Plasmid in COS-7 Cells.
Plasmids pPROTA and pPROTA/β-1,4-GalT II were transiently transfected into COS-7 cells by electroporation (29). COS-7 cells were cultured in DMEM containing 10% FCS for 24 h and then in DMEM containing transferrin (5 μg/ml), insulin (5 μg/ml), and sodium selenite (5 ng/ml). The culture media were harvested after 70 h and incubated with IgG-Sepharose beads (100 μl slurry for 4 ml media) at 4°C for 16 h. The enzyme-containing beads were subsequently collected by centrifugation, washed 10 times with PBS, and suspended in 100 μl of 0.1 M Mes buffer (pH 7.0) containing 4 mM 5′-AMP.
The full coding sequence (nucleotides 1–1164) of the β-1,4-GalT II cDNA was ligated into expression vector pVL1393 (PharMingen), and plasmid pVL1393/β-1,4-GalT II was cotransfected with BaculoGold DNA (PharMingen) into Sf-9 cells grown at 27°C in Grace’s medium containing 10% FCS. The cells were sonicated in 0.1 M Mes buffer (pH 7.0) containing 4 mM 5′-AMP and 1.25% Triton X-100, and the cell homogenates were used for assay of lactose synthetase activity.
Glycosyltransferase Assay and Product Characterization.
Glycosyltransferase assays were performed by incubating 0.1 M Mes buffer (pH 7.0) containing 4 mM 5′-AMP, 250 μM UDP-[3H]sugar (6.64 Ci/mol), 1 mM GlcNAcβ-S-pNP or acceptor as indicated, 20 mM MnCl2, and beads-associated enzyme in a total volume of 50 μl. After incubation at 37°C, the product was isolated by using Sep-Pak C18 cartridges (Waters) and radioactivity incorporated into the product was determined (30). The 3H-labeled products obtained from the acceptors were subjected to digestion with diplococcal β-galactosidase, which cleaves the Galβ1 → 4GlcNAc linkage but not the Galβ1 → 3GlcNAc linkage (31). The galactosylated GlcNAcβ1 → 2Manα1 → 3Manβ1 → O(CH2)8COOCH3 was subjected to RCA-I-agarose column chromatography (32), and the bound fraction was characterized by methylation analysis (33). Lactose synthesis in the presence of α-lactalbumin and lacto-N-neotetraosylceramide synthesis by protein A/β-1,4-GalT II fusion protein were assayed as described previously (14, 15, 34–36). Bovine β-1,4-GalT I was prepared from bovine mammary gland as described previously (37) and incubated with the acceptors listed above.
RESULTS
Isolation of a Human cDNA Related to β-1,4-GalT.
By 3′-RACE of human breast tumor cell line mRNA several PCR fragments were obtained as was revealed by agarose gel electrophoresis. One of the fragments (1,000 bp) most likely was derived from the known human β-1,4-GalT transcript, because restriction fragment analysis revealed a pattern expected for the 3′ part of this cDNA (20). A smaller 860-bp fragment was cloned and sequenced. This PCR fragment showed 39% identity at the amino acid level with human β-1,4-GalT. To isolate a full-length human cDNA clone, a cDNA library was constructed from MRK-nu-1 cells. In a plaque hybridization assay this cDNA library was screened with the α-32P-labeled PCR fragment as a probe. This resulted in the isolation of a cDNA of approximately 4.7 kb. Determination of the nucleotide sequence revealed that the cDNA consists of a 112-bp G/C-rich 5′ noncoding region, followed by an ORF of 1,164 bp and a 3.4-kb long 3′ noncoding region (Fig. 1). The deduced amino acid sequence of the ORF predicts a polypeptide of 388 aa with eight potential N-glycosylation sites (Fig. 1). A hydropathy plot of the amino acid sequence (38) revealed one hydrophobic segment located at amino acid positions 28–48, corresponding to a potential transmembrane domain, suggesting a type-II membrane topology of the polypeptide as typically found in glycosyltransferases (39). Comparison of the deduced amino acid sequence with those of other glycosyltransferases revealed that it has 37% identity with human β-1,4-GalT (ref. 20, Fig. 2) and other mammalian β-1,4-GalTs, and 28% identity with L. stagnalis β-1,4-GlcNAcT (18). Four cysteine residues occur at highly conserved positions in both the protein and human β-1,4-GalT (indicated by solid circles in Fig. 2). In β-1,4-GalT, two of these four cysteine residues (amino acids 129 and 246, respectively) have been reported to form a disulfide bond that is considered to be a prerequisite for the enzyme to interact with α-lactalbumin (40). These results indicate that the cDNA encodes a protein named β-1,4-GalT II related to the mammalian β-1,4-GalT described previously.
Figure 1.
Nucleotide sequence of the 5′-terminal part of β-1,4-GalT II cDNA and its deduced amino acid sequence. The putative transmembrane domain is underlined. The potential N-glycosylation sites are indicated by asterisks. This nucleotide sequence has been submitted to the DDBJ/GenBank/EMBL Data Bank with accession no. AB004550.
Figure 2.
Comparison of the deduced amino acid sequences of human β-1,4-GalT I and β-1,4-GalT II. Gaps introduced to optimize the alignment are indicated by bars. Identical amino acid residues in the sequence alignment are boxed. The putative transmembrane domains are underlined. Cysteine residues are indicated with solid circles.
Expression of a Protein A/β-1,4-GalT II Fusion Plasmid and Characterization of the Secreted Protein.
To obtain evidence that the isolated cDNA encodes a glycosyltransferase, a hybrid construct consisting of part of protein A (28) fused to the stem region of β-1,4-GalT II was transiently transfected to COS-7 cells. This was expected to result in the secretion of an enzymatically active soluble fusion protein (PROTA/β-1,4-GalT II), in the culture medium, that is similar to what has been observed previously with other glycosyltransferases (41–43). The soluble fusion protein was isolated from the medium by using IgG-Sepharose beads, which bind to the protein A part of the fusion protein. The activity of bead-associated fusion protein was assayed by using different sugar-donor and sugar-acceptor substrates. In standard enzyme assays, when using different UDP-sugar donors and GlcNAcβ-S-pNP as an acceptor, PROTA/β-1,4-GalT II showed a GalT activity, whereas no enzyme activity could be detected when using UDP-GalNAc or UDP-GlcNAc as a sugar donor (Table 1). This GalT activity was stimulated in the presence of 20 mM MnCl2, and optimal activity was obtained at pH 7.0 (data not shown). When an IgG-bead suspension that was incubated with pPROTA-transfected COS-7 cell medium was assayed, no detectable activity to the acceptor, regardless of the type of sugar donor used, was found with these beads (Table 1).
Table 1.
Glycosyltransferase activity in the PROTA/β-1,4-GalT II fusion protein produced by COS-7 cells
| Transfected plasmid | Glycosyltransferase activity, pmol/min·mg protein
|
||
|---|---|---|---|
| GalT | GalNAcT | GlcNAcT | |
| pPROTA/β-1,4-GalT II | 302.3 ± 16.9 | 0.5 | 0.2 |
| pPROTA (mock) | 0.1 | 0.1 | 0.1 |
The values represent the averages with standard errors obtained by three independent assays. GalT, GalNAcT, and GlcNAcT indicate galactosyltransferase, N-acetylgalactosaminyltransferase, and N-acetylglucosaminyltransferase, respectively.
To characterize the product obtained with PROTA/β-1,4-GalT II, [3H]galactosylated GlcNAcβ-S-pNP was digested with diplococcal β-galactosidase, which specifically cleaves the Galβ1 → 4GlcNAc linkage but not the Galβ1 → 3GlcNAc linkage (31). After Sep-Pak C18 column chromatography, all the radioactivity was recovered in the pass-through fraction of the column (data not shown), indicating that the galactose was cleaved off efficiently. Galactosylated GlcNAcβ1 → 2Manα1 → 3Manβ1 → O(CH2)8COOCH2-mNP was subjected to RCA-I-agarose column chromatography. RCA-I binds with high affinity to oligosaccharides terminating with β-1,4-linked galactose and binds poorly to those terminating with β-1,3-linked galactose (32, 44). The bound fraction subsequently was subjected to methylation analysis, which showed the presence of 2,3,4,6-tetra-O-methyl galactitol and 3,6-di-O-methyl 2-N-methylacetamido-2-deoxyglucitol at a similar ratio in addition to 3,4,6-tri-O-methyl and 2,4,6-tri-O-methyl mannitols (data not shown). These results indicate that the PROTA/β-1,4-GalT II fusion protein catalyzes transfer of galactose from UDP-Gal to the acceptor in β-1,4-linkage.
GlcNAcβ1 → 6(GlcNAcβ1 → 2)Manα1 → 3Manβ1 → O(CH2)8COOCH2-mNP containing terminal β1 → 6- and/or β1 → 2-linked N-acetylglucosamine could be galactosylated as effectively as GlcNAcβ-S-pNP. However, the galactosylation of GlcNAcβ1 → 2Manα1 → 3Manβ1 → O(CH2)8COOCH2-mNP and GlcNAcβ1 → 4(GlcNAcβ1 → 2)Manα1 → 3Manβ1 → O(CH2)8COOCH2-mNP was less efficient (Table 2). In contrast, little enzyme activity was found by using Galβ-pNP, N-acetylglucosamine, AsAg-human transferrin, or lacto-N-triaosylceramide (data not shown). These results indicate that PROTA/β-1,4-GalT II is able to galactosylate some specific acceptors that have a terminal β-linked N-acetylglucosamine. Because β-1,4-GalT I is also involved in lactose synthesis (15), it was investigated whether α-lactalbumin was able to induce PROTA/β-1,4-GalT II to act on glucose. However, no lactose synthetase activity was observed in the presence of α-lactalbumin (0.5–5 mg/ml).
Table 2.
Partial acceptor specificity of β-1,4-GalT II
| Acceptor | Relative β-1,4-GalT II activity, % |
|---|---|
| 1. GlcNAcβ-S-pNP | 100* |
| 2. GlcNAc | 7 |
| 3. GlcNAcβ1 → 2Manα1 → 3Manβ1 → R† | 26 |
| 4. GlcNAcβ1 ↘ 4Manα1 → 3Manβ1 → R | 34 |
| GlcNAcβ1 ↗ 2 | |
| 5. GlcNAcβ1 ↘ 6Manα1 → 3Manβ1 → R | 109 |
| GlcNAcβ1 ↗ 2 |
The activity of the β-1,4-GalT II was determined at 1 mM of sugar acceptor.
One hundred percent activity corresponds to 274.5 pmol/min·mg protein toward 1 mM GlcNAcβ-S-pNP.
R: O(CH2)8COOCH2-mNP.
The reason why PROTA/β-1,4-GalT II has no lactose synthetase activity could be because of the presence of part of the protein A sequence. To investigate this, the recombinant β-1,4-GalT II with the full coding sequence was expressed by transfection of pVL1393/β-1,4-GalT II in Sf-9 cells that showed little β-1,4-GalT activity toward GlcNAcβ-S-pNP and GlcNAc. Assay of lactose synthetase activity when using the cell homogenates as an enzyme source revealed that the gene-transfected cells, which acquired a high level of β-1,4-GalT activity to GlcNAcβ-S-pNP but not GlcNAc, contain no activity toward glucose in the presence of increasing concentrations of α-lactalbumin (unpublished data). These results showed that the presence of part of the protein A sequence in PROTA/β-1,4-GalT II is not responsible for the lack of lactose synthetase activity because the recombinant protein with only the full coding sequence also did not have such activity. Thus, β-1,4-GalT II is a transferase that has no lactose synthetase activity.
Expression of β-1,4-GalT II mRNA in Human Tissues.
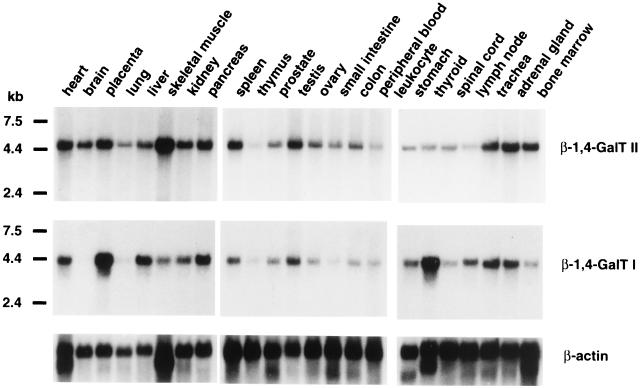
The expression of β-1,4-GalT II mRNA in a variety of human tissues was examined by Northern blot analysis. Commercially available Northern blots containing poly(A)+ RNA from human tissues were hybridized with β-1,4-GalT II cDNA as a probe. A single transcript of approximately 4.7 kb was detected in all 24 tissues examined, although the levels of the transcript varied among different tissues (Fig. 3 Top). When the same blots were hybridized with human β-1,4-GalT I cDNA as a probe, a single transcript of 4.2 kb was detected in most tissues except brain and lung (Fig. 3 Bottom). Comparison of the 4.7-kb and 4.2-kb transcript levels revealed that relatively high levels of the β-1,4-GalT II message are found in brain, lung, skeletal muscle, and bone marrow, in which no detectable or low amounts of the β-1,4-GalT I message are observed. In thyroid, more β-1,4-GalT I mRNA was observed than β-1,4-GalT II mRNA, whereas in spleen and bone marrow, more β-1,4-GalT II mRNA was observed than β-1,4-GalT I mRNA. Thus, the β-1,4-GalT II gene and β-1,4-GalT I gene are both widely expressed in human tissues but at different relative levels.
Figure 3.
Northern blot analysis of β-1,4-GalT II and β-1,4-GalT I transcripts in human tissues. Commercially available mRNA blots containing 2 μg of poly(A)+ RNA/lane were probed with 32P-labeled β-1,4-GalT II cDNA, human β-1,4-GalT I cDNA, or β-actin cDNA. The sizes of RNA markers are indicated in kilobases (kb).
DISCUSSION
In the present study we have described the isolation of a cDNA clone encoding a novel β-1,4-GalT from a cDNA library of the human breast tumor cell line, MRK-nu-1. The deduced amino acid sequence of this cDNA shows 37% identity with that of the previously described human β-1,4-GalT cDNA. To discriminate these transferases, we named the present β-1,4-GalT as β-1,4-GalT II whereas we refer to the well described β-1,4-GalT as β-1,4-GalT I in the present paper. The isolated 4.7-kb cDNA most likely represents a full-length cDNA because Northern blot analysis revealed a single transcript of the similar size. Hydropathy plot analysis of the deduced amino acid sequence indicated a hydrophobic segment (amino acids 28–48) that may be a transmembrane domain. However, topology refinement based on the helical transmembrane structures (45) revealed alternatively that the region containing amino acids 17–33 could represent that domain. In either case, however, the putative transmembrane domain is located near the N terminus of the protein, indicating a type-II membrane topology that is typical for glycosyltransferases.
The cloning strategy employed in this study was based on the occurrence of several highly conserved regions in the putative catalytic domains of mammalian β-1,4-GalTs and L. stagnalis β-1,4-GlcNAcT (18, 24). In particular, a highly conserved region of 10 aa, the (Y/F)(W/F)GWG(G/A)EDD(D/E) box, has been very useful in this study. By 3′-RACE with degenerate primers based on this box, a 860-bp PCR fragment was obtained from human breast tumor cell line mRNA, which subsequently was used as a probe to isolate a human cDNA. Indeed, the cloned β-1,4-GalT II cDNA shows most of the conserved regions as described above, allowing its identification as a novel member of the β-1,4-GalT gene family (24). The 3′-RACE used thus has proven to be a powerful technique to isolate novel members of this gene family. Related cDNA fragments have been obtained from mRNA preparations derived from bovine mammary gland and mouse brain (unpublished data) and also have been reported as expressed sequence tags (accession nos. AA013728 and AA015244), suggesting the existence of a similar gene in cow and mouse. These data suggest that the β-1,4-GalT I and β-1,4-GalT II genes form separate lineages in mammals and may have evolved from a common ancestor gene by duplication event that happened before radiation of mouse, cow, and man.
Initial acceptor specificity studies suggest that differences may be expected between the two enzymes in their affinity for oligosaccharide substrates because the sequence identity between them is only 37%. Quite interestingly, β-1,4-GalT II seems to galactosylate acceptors containing GlcNAcβ1 → 6(GlcNAcβ1 → 2)Man structure more efficiently than those containing GlcNAcβ1 → 2Man and GlcNAcβ1 → 4(GlcNAcβ1 → 2)Man structures. These results suggest that β-1,4-GalT II prefers terminal N-acetylglucosamine residue, which is β1 → 6 linked as found in tri- and tetraantennary complex-type oligosaccharides. This property is also found in the genetically related L. stagnalis β-1,4-GlcNAcT (19). A marked difference was observed in lactose synthesis because the novel β-1,4-GalT showed no ability to transfer galactose from UDP-Gal to glucose in the presence of α-lactalbumin. Under the same conditions bovine mammary gland β-1,4-GalT I synthesized lactose. In addition, β-1,4-GalT II may not be involved in lacto-series glycolipid synthesis because no appreciable transfer of galactose was observed to lacto-N-triaosylceramide. Further studies on the acceptor specificity as well as kinetics of this novel β-1,4-GalT, however, are necessary to get more insight in the specific biological function of this enzyme.
Northern blot analysis revealed that β-1,4-GalT II is expressed in most human tissues like β-1,4-GalT I, although the relative expression levels of the transcript vary. This raises the question as to the specific in vivo function of either enzyme. The β-1,4-GalT I has been proposed to have a housekeeping function in addition to a specific function in the mammary gland, where it is involved in lactose synthesis on interaction with α-lactalbumin (9, 15). In β-1,4-GalT I-deficient mice, there was residual galactosylation of N-linked oligosaccharides, which indeed might result from the action of β-1,4-GalT II. However, the milk from such mutant mice does not contain lactose (13), which is consistent with the unresponsiveness of the latter enzyme to α-lactalbumin. Remarkably, in human brain a significant level of β-1,4-GalT II transcript was present whereas no β-1,4-GalT I message was found. Similarly, mouse brain has been reported to express low levels of β-1,4-GalT I transcript (46). Because mouse brain glycoproteins have been reported to contain N-linked oligosaccharides with Galβ1 → 4GlcNAc groups that may carry the HNK-1 epitope or polysialic acid (4), it might be expected that β-1,4-GalT II is the enzyme that is involved in the galactosylation of brain glycoproteins rather than β-1,4-GalT I. In other tissues both enzymes may participate in the cooperative galactosylation of different substrates in a way that is, so far, unknown. This might be important, for example, in the case of B cells producing IgG molecules, the galactosylation of which can vary over the course of rheumatoid arthritis and other inflammatory diseases (reviewed in refs. 47 and 48). In this context it is of interest to note that β-1,4-GalT II shows considerable expression in bone marrow, a tissue that is rich in B cells. Furthermore, in the lung of β-1,4-GalT I-deficient mice, in which no transcript nor activity of the transferase could be detected, high levels of β-1,4-galactosylated proteins were found (12). This suggests that these proteins can be β-1,4-galactosylated in an alternative pathway. In human lung, much higher β-1,4-GalT II transcript levels than β-1,4-GalT I transcript levels are observed. A similar expression pattern in mouse lung would account for the presence of such β-1,4-linked galactose on the glycoproteins in this tissue. The discovery of this novel β-1,4-GalT will lead to further understanding of the biological significance of protein galactosylation.
Acknowledgments
We thank Dr. Michiko Fukuda at the Burnham Institute (La Jolla, CA) for providing us a human β-1,4-GalT I cDNA clone. This work was supported by the Grants-in-Aid for Scientific Research on Priority Areas No. 05274106 and No. 09240104 from the Ministry of Education, Science, Culture, and Sport of Japan.
Note Added in Proof
During processing of this manuscript, related but not identical human β-1,4-GalTs have been described by Almeida, R., et al. (49). These β-1,4-GalTs have been named β-4Gal-T2 and β-4Gal-T3. Therefore, we propose to refer to β-1,4-GalT II described here as β-1,4-GalT IV in the future.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: RACE, rapid amplification of cDNA ends.
Data deposition: The nucleotide sequence reported in this paper has been deposited in the DDBJ/GenBank/EMBL database (accession no. AB004550).
References
- 1.Hakomori S. Curr Opin Immunol. 1991;3:646–653. doi: 10.1016/0952-7915(91)90091-e. [DOI] [PubMed] [Google Scholar]
- 2.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhavanadan V P, Furukawa K. In: Biology of the Sialic Acids. Rosenberg A, editor. New York: Plenum; 1995. pp. 145–196. [Google Scholar]
- 4.Kunemund V, Jungalwala F B, Fischer G, Chou D K H, Keilhauer G, Schachner M. J Cell Biol. 1988;106:213–223. doi: 10.1083/jcb.106.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schachter H. Glycobiology. 1991;1:453–461. doi: 10.1093/glycob/1.5.453. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar M, Hull E, Nishikawa Y, Simpson R J, Moritz R L, Dunn R, Schachter H. Proc Natl Acad Sci USA. 1991;88:234–238. doi: 10.1073/pnas.88.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brew K, Vanaman T C, Hill R L. Proc Natl Acad Sci USA. 1968;59:491–497. doi: 10.1073/pnas.59.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Agostaro G, Bendiak B, Tropak M. Eur J Biochem. 1989;183:211–217. doi: 10.1111/j.1432-1033.1989.tb14915.x. [DOI] [PubMed] [Google Scholar]
- 9.Rajput B, Shaper N L, Shaper J H. J Biol Chem. 1996;271:5131–5142. doi: 10.1074/jbc.271.9.5131. [DOI] [PubMed] [Google Scholar]
- 10.Ioffe E, Stanley P. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzler M, Gertz A, Sarkar M, Schachter H, Schrader J W, Marth J D. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Q, Hasty P, Shur B D. Dev Biol. 1997;181:257–267. doi: 10.1006/dbio.1996.8444. [DOI] [PubMed] [Google Scholar]
- 13.Asano M, Furukawa K, Kido M, Matsumoto S, Umesaki Y, Kochibe N, Iwakura Y. EMBO J. 1997;16:1850–1857. doi: 10.1093/emboj/16.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison J F, Ebner K E. J Biol Chem. 1971;246:3992–3998. [PubMed] [Google Scholar]
- 15.Hill R L, Brew K. Adv Enzymol Rel Areas Mol Biol. 1975;43:411–490. doi: 10.1002/9780470122884.ch5. [DOI] [PubMed] [Google Scholar]
- 16.Sheares B, T, Carlson D M. J Biol Chem. 1984;259:8045–8047. [PubMed] [Google Scholar]
- 17.Rivera-Marrero C A, Cummings R D. Mol Biochem Parasitol. 1990;43:59–68. doi: 10.1016/0166-6851(90)90130-e. [DOI] [PubMed] [Google Scholar]
- 18.Bakker H, Agterberg M, Van Tetering A, Koeleman C A M, Van den Eijnden D H, Van Die I. J Biol Chem. 1994;269:30326–30333. [PubMed] [Google Scholar]
- 19.Bakker H, Schoenmakers P S, Koeleman C A M, Joziasse D H, Van Die I, Van den Eijnden D H. Glycobiology. 1997;7:539–548. doi: 10.1093/glycob/7.4.539. [DOI] [PubMed] [Google Scholar]
- 20.Masri K A, Appert H E, Fukuda M N. Biochem Biophys Res Comm. 1988;157:657–663. doi: 10.1016/s0006-291x(88)80300-0. [DOI] [PubMed] [Google Scholar]
- 21.Shaper N L, Hollis G F, Douglas J G, Kirsch I R, Shaper J H. J Biol Chem. 1988;263:10420–10428. [PubMed] [Google Scholar]
- 22.Shaper J H, Joziasse D H, Meurer J A, Chou T D, Schnaar R L, Shaper N L. Glycoconjugate J. 1995;12:477–478. [Google Scholar]
- 23.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 24.Van Die I, Bakker H, Van den Eijnden D H. Glycobiology. 1997;7:v–ix. doi: 10.1093/glycob/7.8.1053. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa K, Matsuta K, Takeuchi F, Kosuge E, Miyamoto T, Kobata A. Intern Immunol. 1990;2:105–112. doi: 10.1093/intimm/2.1.105. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 28.Sanchez-Lopez R, Nicholson R, Gesnel M-C, Matrisian L M, Breathnach R. J Biol Chem. 1988;263:11892–11899. [PubMed] [Google Scholar]
- 29.Chu G, Hayakawa H, Berg P. Nucleic Acids Res. 1987;15:1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palcic M M, Heerze L D, Pierce M, Hindsgaul O. Glycoconjugate J. 1990;5:49–63. [Google Scholar]
- 31.Glasgow L R, Paulson J C, Hill R L. J Biol Chem. 1977;252:8615–8623. [PubMed] [Google Scholar]
- 32.Merkle R K, Cummings R D. Methods Enzymol. 1987;138:232–259. doi: 10.1016/0076-6879(87)38020-6. [DOI] [PubMed] [Google Scholar]
- 33.Furukawa K, Roberts D D, Endo T, Kobata A. Arch Biochem Biophys. 1989;270:302–312. doi: 10.1016/0003-9861(89)90032-5. [DOI] [PubMed] [Google Scholar]
- 34.Basu M, De T, Das K K, Kyle J W, Chon H-C, Schaeper R J, Basu S. Methods Enzymol. 1987;138:575–607. doi: 10.1016/0076-6879(87)38053-x. [DOI] [PubMed] [Google Scholar]
- 35.Ebner K E, Mawal R, Fitzgerald D K, Colvin B. Methods Enzymol. 1972;28B:507–510. [Google Scholar]
- 36.Nakazawa K, Furukawa K, Narimatsu H, Kobata A. J Biochem. 1993;113:747–753. doi: 10.1093/oxfordjournals.jbchem.a124115. [DOI] [PubMed] [Google Scholar]
- 37.Furukawa K, Roth S. Biochem J. 1985;227:573–582. doi: 10.1042/bj2270573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 39.Paulson J C, Colley K J. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 40.Yadav S P, Brew K. J Biol Chem. 1991;266:698–703. [PubMed] [Google Scholar]
- 41.Kurosawa N, Inoue M, Yoshida Y, Tsuji S. J Biol Chem. 1996;271:15109–15116. doi: 10.1074/jbc.271.25.15109. [DOI] [PubMed] [Google Scholar]
- 42.Bierhuizen M A, Fukuda M. Proc Natl Acad Sci USA. 1992;89:9326–9330. doi: 10.1073/pnas.89.19.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakker H, Van Tetering A, Agterberg M, Smit A B, Van den Eijnden D H, Van Die I. J Biol Chem. 1997;272:18580–18585. doi: 10.1074/jbc.272.30.18580. [DOI] [PubMed] [Google Scholar]
- 44.Baenziger J U, Fiete D. J Biol Chem. 1979;254:9795–9799. [PubMed] [Google Scholar]
- 45.Rost B, Casadio R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harduin-Lepers A, Shaper J H, Shaper N L. J Biol Chem. 1993;268:14348–14359. [PubMed] [Google Scholar]
- 47.Furukawa K, Kobata A. Mol Immunol. 1991;28:1333–1340. doi: 10.1016/0161-5890(91)90035-i. [DOI] [PubMed] [Google Scholar]
- 48.Filley E, Andreoli A, Steele J, Waters M, Wagner D, Nelson D, Tung K, Rademacher T, Dwek R, Rook G A W. Clin Exp Immunol. 1989;76:343–347. [PMC free article] [PubMed] [Google Scholar]
- 49.Almeida R, Amado M, David L, Levery S B, Holmes E H, Merkx G, van Kessel A G, Rygaard E, Hassan H, Bennett E, Clausen H. J Biol Chem. 1997;272:31979–31991. doi: 10.1074/jbc.272.51.31979. [DOI] [PubMed] [Google Scholar]