Abstract
Chaperonin GroEL has been found to interact with isolated cytoplasmic membrane of Escherichia coli. Interaction requires Mg ions, whereas MgATP inhibits, and inhibition is stronger in the presence of co-chaperonin GroES. “Heat-shock” of the membrane at 45°C destroys irreversibly its ability to bind GroEL. The binding of GroEL is characterized by saturation with a maximum of about 100 pmol GroEL bound per mg of total membrane protein, indicating a limited capacity and specificity of the membrane to bind GroEL. According to results of immunoblotting analysis and cleavable photoactivable cross-linking, a membrane target of GroEL is SecA, a protein known as a central component of the translocation machinery. Moreover, in some cases GroEL could modulate a cycle of association of SecA with the membrane by stimulating release of SecA from the membrane. A physiological role of targeting of GroEL in or close to the protein-conducting membrane apparatus is discussed.
Keywords: heat shock, SecA51Tsmutant, photo-cross-linking
The Escherichia coli heat-shock protein GroEL, which is a double toroid comprising 14 identical subunits of 57.3 kDa, belongs to the highly conserved Hsp60 (cpn60) family of molecular chaperones named chaperonins. As a molecular chaperone, GroEL is required for correct folding and assembly of newly synthesized proteins and for recovery of the cell after exposure to either thermal or chemical stress, which denatures protein components (1–3). GroEL interacts with nonfolded newly synthesized proteins in an MgATP-dependent fashion (4) and promotes their folding presumably either by preventing or reversing unspecific aggregation and other unfavorable interactions within and between polypeptide chains. Numerous experiments suggest that the GroEL–substrate protein complexes are maintained through hydrophobic contacts. The binding and hydrolysis of ATP by GroEL are crucial for its functioning and are assumed to be necessary for the binding–release cycles of substrate proteins, which drive the substrate folding. For the folding or assembly of some proteins, the MgATP (or MgADP)-dependent specific interaction of GroEL with its helper protein GroES (7 × 10 kDa) is also required. Whether proteins fold to native structure while they are bound to GroEL or whether they have to be released into solution to complete folding is unclear.
Apart from its recognized function in protein folding, GroEL is able to play a role in protein translocation across (4–7) or insertion into (8, 9) the cytoplasmic membrane of E. coli. Export of some secretory proteins are damaged in E. coli strains functionally defective in GroEL (10, 11). The temperature-sensitive phenotype of several mutant proteins are suppressed by overexpression of GroEL (12–14). Interestingly, some of these mutants are the membrane proteins SecA and SecY, which are key components of the preprotein translocation apparatus in E. coli and are presumed to form a protein-conducting membrane channel (15). All of this evidence from genetic and biochemical studies, in vivo and in vitro, allows us to suggest that GroEL not only is able to maintain newly synthesized secretory or membrane proteins in a state competent for interaction with membrane but it also could play a direct role in the membrane targeting of these proteins. To investigate this question, we found that purified GroEL interacts with the isolated cytoplasmic membrane of E. coli and, specifically, with SecA associated with the membrane. Some important features and physiological consequences of this interaction are discussed.
MATERIALS AND METHODS
Bacterial Strains and Preparations.
E. coli strains MC4100 or MM52 carrying the wild-type (wt)-SecA or SecA51(Ts) mutant, respectively, were grown aerobically at 30°C in Luria–Bertani (LB) medium to OD600 ≈ 1.0. E. coli strain T184 (16) was cotransformed with pGP1–2 harboring gene of T7 RNA polymerase (17) and pT7SecA2 carrying gene of SecA under T7 promoter (18). Purification of chaperonins GroEL and GroES were described by us previously (19). Molar concentrations of GroEL and GroES are expressed in terms of their particles (14-mer and 7-mer, respectively). Rabbit affinity-purified GroEL IgG antibodies were prepared according to ref. 4.
Isolation of IMVs.
E. coli MC4100 or MM52 cells were disrupted by sonicating (Microson, Heat Systems/Ultrasonics) in buffer containing 25 mM KCl/1 mM MgCl2/0.2 mM EDTA/1 mM DTT/0.5 mM PMSF/10 mM Tris⋅HCl/25 mM TEA⋅HCl, pH 7.5 (20). The homogenate was clarified, and crude IMVs were isolated by centrifugation at 60,000 rpm for 75 min at 4°C (TLA100.1 rotor, Beckman TL100 centrifuge). The pelleted IMVs were suspended in 7.5% sucrose/1 mM MgCl2/0.1 mM EDTA/1 mM DTT/50 mM triethanolamine-OAc, pH 7.5, and purified by recentrifugation through 22% sucrose in 50 mM KCl/20 mM MgCl2/0.5 mM EDTA/1 mM DTT/30 mM TEA-OAc, pH 7.5, at 54,000 rpm for 90 min at 4°C (TLS55 rotor, Beckman TL100 centrifuge). Where indicated (Fig. 2), purified cytoplasmic inverted cytoplasmic membrane vesicles (IMVs) were isolated by passing cell suspension twice through a French pressure cell, followed by separation of inner membrane vesicles from outer membrane by centrifugation through sucrose gradient containing 20 mM EDTA (21). As described (22), the presence of EDTA did not interfere with interaction of IMVs with SecA and was used for isolation of IMV–SecA complex.
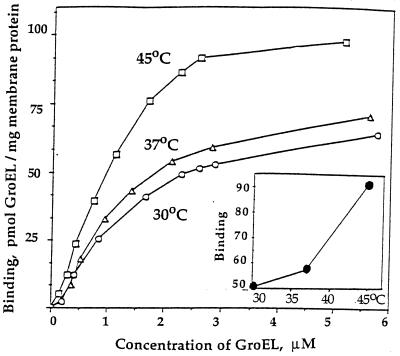
Figure 2.
Effects of concentration of GroEL on its interaction with IMVs at different temperatures. The purified cytoplasmic IMVs were used. (Inset) The temperature dependence of binding of GroEL to IMVs (at 2.5 μM GroEL).
The [35S]IMVs containing radioactive wt-SecA were isolated from T184 (pGP1–2, pT7SecA2) cells. The cells were grown at 30°C in LB medium (plus 30 μg/ml kanamycin and 100 μg/ml ampicillin) to OD600 ≈ 0.6 and additionally for 1 hr in M9 minimal medium supplemented with 0.5% glucose and 40 μg/ml amino acids (lacking methionine and cysteine). After methionine starvation, the culture was incubated for 20 min at 42°C (induction of T7 RNA polymerase) and for an additional 20 min in the presence of 0.4 mM rifampicin. The cells were cooled to 30°C, and growth was continued for 30 min in the presence of ≈20 μCi/ml of l-[35S]Met (specific activity, >1,000 Ci/mmol, Amersham). The cells were then harvested on ice and washed with 10% sucrose/10 mM Tris⋅HCl, pH 8.0.
Interaction of GroEL with IMVs.
Typically, interaction of GroEL with IMVs was carried out in a 40-μl sample containing 40–70 μg IMVs and 1 μM GroEL in 10 mM MgCl2/0.1 mM EDTA/25 mM KCl/5 mM DTT/50 mM TEA-OAc, pH 7.5 (“binding buffer”). The samples were incubated for 20 min at different temperatures and loaded on a four-step sucrose gradient (0.34 ml each of 20, 15, 10, and 7.5% in the binding buffer). Where indicated, some additions to the incubation mixtures and sucrose gradients were included. After centrifugation at 4°C for 70 min at 54,000 rpm (TLS55 rotor, Beckman TL100 centrifuge), 120-μl fractions were collected from the top. The membrane pellets dissolved in 33 μl (final volume) 2% SDS-sample buffer for 30 min at 37°C were analyzed by SDS/PAGE by using 8% acrylamide/0.16% bisacrylamide. After Coomassie blue staining, the GroEL bands were quantitated by scanning densitometry relative to a known GroEL standards loaded on the same gel (Model GS-690 Imaging Densitometer, Bio-Rad). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose membrane and immunoblotting with SecA antisera were performed according to enhanced chemiluminescence (ECL) Western blotting protocol (Amersham) as described by the manufacturer.
Cross-Linking of GroEL to SecA on the Membrane.
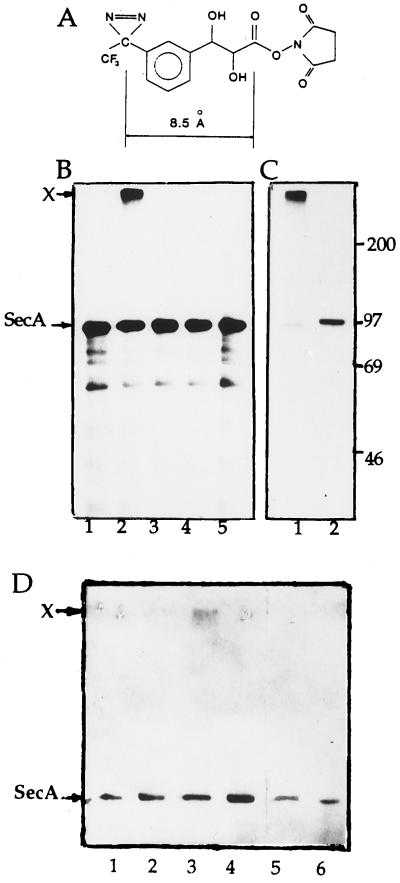
The coupling of the photoactivable cross-linker TDPDP (23) to the amino groups of GroEL was carried out by treatment of 3.75 μM GroEL (i.e., ≈2.1 mM amino groups) with 2.8 mM 3-{3-[3-(trifluoromethyl)diazirin-3-yl]phenyl}-2,3-dihydroxypropionic acid N-hydroxysuccinimide ester (TDPDP-ONSu (Fig. 4A) for 1 hr at 25°C in the binding buffer, except that 0.4 mM DTT and 10% dimethyl sulfoxide were presented. The reaction was stopped by treatment for 15 min at 25°C with 1/10 volume of 1 M Tris⋅HCl, pH 8.0, and the photoactivable TDPDP-GroEL was separated from the reagent excess by gel filtration in the same buffer. According to a test with a colorimetric amino-group reagent TNBS (24), GroEL particle acquired ≈4 photoactivable groups per subunit. TDPDP-GroEL (0.5 μM) was incubated for 20 min at 37°C with 37 μg [35S]IMVs (≈2 × 106 cpm) in 40 μl binding buffer containing, where indicated, some additions (Fig. 4A). Irradiation was performed for 1 min on ice with a UVP 350-W mercury short-arc lamp with a filter transmitting light preferentially above 330 nm and 2-cm water layer to prevent heating of the sample (conditions of complete decomposition of the photo-probe).
Figure 4.
Photo-induced cross-linking of the TDPDP-GroEL to SecA on the membranes. (A) Structure of cross-linker TDPDP-ONSu (23). (B) Cross-linking of the TDPDP-GroEL to [35S]IMVs is caused by targeting of GroEL to the membrane. Lanes: 1, GroEL and [35S]IMVs in the presence of 10 mM MgCl2, no irradiation; 2, as in lane 1 but after irradiation; 3, as in lane 2, except that 10 mM MgCl2 is replaced by 20 mM EDTA; 4, as in lane 2 but in the presence of 7.5 mM ATP plus 1.4 μM GroES; 5, [35S]IMVs irradiated alone. (C) 35S-labeled membrane component cross-linked to GroEL is indeed the SecA. The sample identical to that shown in lane 2 of B was used, and GroEL carrying the radioactive label was isolated by treatment of the membranes with 0.8% DM followed by centrifugation in the sucrose gradient, as described in Materials and Methods, and analyzed by SDS/PAGE and fluorography before or after periodate treatment (lanes 1 or 2, respectively). Band X, cross-linked product. Standard molecular mass markers (Amersham) are indicated on the right (in kDa). (D) Immunoblotting detection with SecA antisera of cross-linking of TDPDP-GroEL to the chromosomally produced wt-SecA on the membrane. Immunoblots of the GroEL-containing fractions (isolated by centrifugation of the IMVs after irradiation in the presence of TDPDP-GroEL and solubilization by DM as described above) before and after periodate treatment are shown in odd or even lanes, respectively. Lanes: 1 and 2, GroEL and IMVs in the presence of 10 mM MgCl2, no irradiation; 3 and 4, as in lanes 1 and 2 but after irradiation; 5 and 6, as in lanes 3 and 4, except that 10 mM MgCl2 is replaced by 20 mM EDTA. Note: A slight intensity of the band X in lane 3 is likely caused by a low yield of electrophoretic transfer of cross-linking product X (>200 kDa) from polyacrylamide gel to nitrocellulose membrane before immunoblotting.
The irradiated [35S]IMVs were sedimented through a sucrose gradient as described in previous paragraph, and membrane pellets were solubilized in 40 μl of 10 mM MgCl2/25 mM KCl/2 mM DTT/0.1 mM EDTA/50 mM Tris⋅HCl, pH 7.5, containing 0.8% nonionic detergent dodecylmaltoside (DM). Ten microliters of the solubilized membranes was analyzed by SDS/PAGE. The remaining 30 μl was centrifuged at 4°C for 130 min at 54,000 rpm (TLS55 rotor, Beckman TL100 centrifuge) through 5–20% sucrose gradient prepared on the same buffer but with 0.2% DM. The gradients were divided into 110-μl fractions from the top. The gradient fractions containing GroEL with the cross-linked [35S] label were combined and precipitated with 5% trichloroacetic acid on ice. The pellet was solubilized for 30 min at 37°C in 25 μl 1.8% SDS/5 mM N-ethylmaleimide/15 mM methylamine/0.1 M sodium phosphate, pH 7.5, and was divided into two 12-μl aliquots. One aliquot was directly analyzed by SDS/PAGE and fluorography whereas the another was treated first for 30 min at 25°C with 1.3 μl 0.1 M sodium metaperiodate (BDH). Then, DTT was added to 20 mM to destroy the excess of periodate, and the sample was analyzed by SDS/PAGE.
The analogous procedure was applied for cross-linking of photoactivable TDPDP-GroEL to IMVs carrying the chromosomally produced wt-SecA. Identification of the SecA in the isolated cross-linking product was carried out by immunoblotting with SecA antisera as described above.
RESULTS
GroEL Interacts with Cytoplasmic Membrane of E. coli.
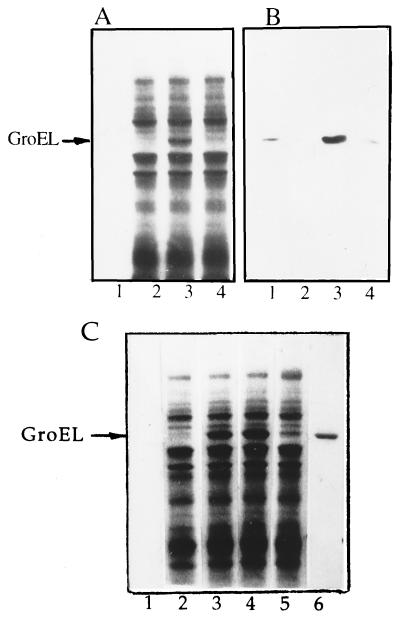
Incubation of GroEL with IMVs followed by isolation of the membranes by sedimentation results in an appearance in the membrane protein profile of a new and well defined Coomassie-stained band (Fig. 1A, lane 3) which, according to immunoblotting analysis with anti-GroEL IgG (Fig. 1B, lane 3), corresponds to GroEL. As seen in Fig. 1 A and B, IMVs themselves are not contaminated by endogenous GroEL (lane 2). Binding of GroEL to IMVs is promoted by Mg ions and is inhibited by EDTA (compare lanes 3 and 4). The inhibitory effect of EDTA is not caused by EDTA-induced removal from IMVs of a special membrane protein(s) required to anchor GroEL. In particular, treatment of IMVs with 20 mM EDTA followed by their sedimentation and resuspension in the Mg-containing buffer does not change the yield of the GroEL–IMV complex (data not shown). Possibly, Mg ions are required to prevent the electrostatic repulsion between negatively charged membrane surface and GroEL (pI ∼ 4.48, ref. 25).
Figure 1.
Interaction of GroEL with IMVs. (A and B) Cosedimentation of GroEL with IMVs after incubation at 37°C in the presence of 10 mM MgCl2 (lanes 1–3) or 20 mM EDTA (lane 4). (A) Coomassie blue R-250 staining. (B) Western blotting using GroEL IgG antibodies. Lanes: 1, GroEL alone; 2, IMVs alone; 3 and 4, GroEL plus IMVs. (C) Heating of IMVs at 45°C inhibits irreversibly their subsequent interaction with GroEL at 30°C (Coomassie staining). Lanes: 1, GroEL alone, incubation for 30 min at 45°C; 2, IMVs alone, incubation for 30 min at 30°C; 3, GroEL plus IMVs, incubation for 30 min at 30°C; 4, GroEL plus IMVs, incubation for 30 min at 45°C and then for 30 min at 30°C; 5, IMVs alone, incubation for 30 min at 45°C and, after cooling to 30°C, GroEL was added and incubation was continued for 30 min at 30°C; 6, GroEL used as a marker.
Table 1 shows the effects of adenine nucleotides and co-chaperonin GroES on interaction of GroEL with IMVs. The presence of 0.1 mM ADP or ATP has no effect whereas addition of co-chaperonin GroES or ATP-regenerating system (to ATP) decreases strongly the yield of the membrane-bound GroEL. At high concentration (3 mM), ADP and especially ATP alone show remarkable inhibitory effects. Sensitivity of GroEL–IMV complex to adenine nucleotides (and GroES) is similar to that of the GroEL complex with various nonnative substrate proteins (3) and is contrary to that of the GroEL interaction with lipids (26). This implies that, upon membrane attachment, GroEL interacts with protein rather then with lipid. Additional evidence in favor of this suggestion comes from measuring GroEL–IMV interaction as a function of concentration of GroEL at different temperatures. The binding of GroEL to IMVs versus concentration of GroEL shows a saturation indicating a restricted capacity of IMVs for binding GroEL (Fig. 2). In particular, at about 2–3 μM GroEL the curves reach a plateau approximately 55–60 or 95–100 pmol GroEL bound per mg total membrane protein at 30–37°C or 45°C, respectively. These values are estimated to be roughly comparable, for example, to concentrations in cytoplasmic membrane of E. coli of each of Sec proteins such as SecA, SecD, SecE, or SecY and about five times less than that for ATP-synthase (22, 27–29). The clear jump in the capacity for the GroEL–IMV interaction at 45°C is probably caused by additional exposure and/or unfolding of the membrane target at this temperature.
Table 1.
Effect of adenine nucleotides and GroES on GroEL binding to IMV at 37°C
| Experiment | Additions | GroEL binding, % |
|---|---|---|
| 1 | — | 100 |
| 2 | 0.1 mM ADP | 99 (66) |
| 3 | 0.1 mM ADP + GroES | 12 |
| 4 | 0.1 mM ATP | 101 (23) |
| 5 | 0.1 mM ATP* | 30 |
| 6 | 0.1 mM ATP + GroES | 15 |
| 7 | 0.1 mM ATP* + GroES | 5 |
Molar ratio, GroES:GroEL = 2.5. Asterisk shows that the ATP-regenerating system (10 mM creatine phosphate and 50 μg/ml creatine kinase) was included. The numbers in parantheses indicate the GroEL binding to IMVs at 3.0 mM ADP or ATP.
Another interesting feature of the curves in Fig. 2 is their sigmoidicity, which is usually interpreted in terms of a positive cooperativity. The slopes of the Hill plots (not shown) are practically independent on the temperatures used, and the average value of the Hill coefficient nH is about 1.7 ± 0.06, suggesting that membrane target for GroEL is perhaps an oligomeric protein, at least a dimer. An additional restriction on the nature of the target is that this protein must be in a partially unfolded state to be recognized by GroEL. From data in Fig. 2 the apparent dissociation constant Kd for GroEL–IMV complex is roughly 1.0 μM, with a slight dependence on temperature. This value of Kd is biologically relevant because it does not exceed the physiological concentration of GroEL (close to 1 μM, ref. 3).
Interaction of IMVs, preheated at 45°C, with GroEL at 30°C is much lower compared with that for the IMVs that are not preheated (Fig. 1C, lanes 5 and 3, respectively). Apparently, cooling of the membrane target denatured at 45°C yields a conformation that is no longer recognized by GroEL, i.e., the temperature-induced unfolding of the membrane target for GroEL is irreversible. Such phenomenon of heat-dependent conformational irreversibility is known to be a typical feature of a variety of proteins and is in contradistinction to the temperature-promoted phase transition of lipids, which is reversible (30). In addition, the phase transition of E. coli cytoplasmic membrane is always completed below growth temperature (31, 32), i.e., below 30°C under our conditions. Considered together with these data, the heat-induced irreversible block of the binding ability of GroEL to the membrane is indicative of conformational damage in membrane protein(s), interacting with GroEL, and not in lipids.
GroEL Interacts with the Membrane-Bound SecA and in Some Cases Could Promote Its Release from the Membrane.
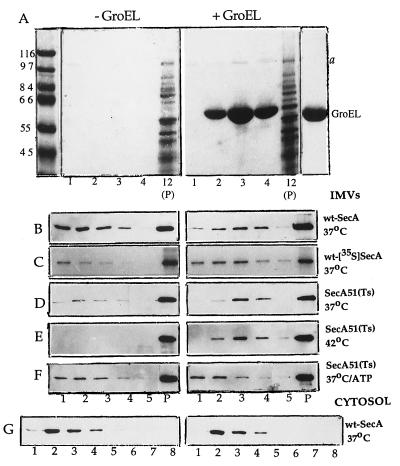
Identification of membrane protein(s) interacting with GroEL is of importance in elucidating the physiological role of interaction of GroEL with the membrane. In this context, a striking finding was observed when IMVs were separated from excess of unbound GroEL by sucrose gradient sedimentation, namely, a faint band a of about 100 kDa, stained by Coomassie blue, which preferentially sediments together with GroEL (Fig. 3A, fraction 3). Because the GroEL preparation itself, overloaded on the same gel, is not contaminated with this band (Fig. 3A, right lane), it seems likely that a protein interacting with GroEL originates from the membrane and, consequently, the protein a-dissociating action of GroEL on the membrane is very specific. The following observations suggest that band a corresponds to the membrane-associated protein SecA. SecA is known to be a cytoplasmically exposed membrane protein (27) of molecular mass of 102 kDa (34), which exists in both soluble and membrane-associated forms (33, 34). That GroEL suppresses the protein translocation defect in the SecA51(Ts) mutant at nonpermissive temperature 42°C (11–13) is also evidence in favor of the possibility that GroEL interacts with SecA. Protein FtsY, which also exists in soluble and membrane-bound states and migrates in SDS gel as an ≈100-kDa polypeptide (35), or some other membrane proteins could be also considered.
Figure 3.
GroEL interacts with SecA associated with the membrane. The IMVs carrying either wt-SecA originated from chromosome (A and B) or wt-[35S]SecA originated from plasmid and metabolically labeled with [35S]methionine (C), or SecA51(Ts) mutant (D–F) were used. In G, the membrane-depleted cellular extract containing soluble fraction of wt-SecA was used instead of wt-IMVs. [Note that a similar picture was observed when the membrane-depleted cellular extract containing Sec51(Ts) mutant was used (not shown)]. The samples were incubated for 30 min at indicated temperatures in the absence or presence of 1.5 μM GroEL in the Mg-containing buffer. In F, 5 mM ATP plus 3.8 μM GroES was also added. The membranes were then sedimented through a sucrose gradient for 70 min at 54,000 rpm as described in Materials and Methods. In G, for better separation of GroEL from soluble wt-SecA, the mixtures were centrifuged for 120 min so that the GroEL maximum was in fractions 6–7 from the top. The fractions were collected from the top of the gradients (total, 11). The corresponding aliquots from the gradient fractions and membrane pellets (P), solubilized in SDS-sample buffer, were analyzed by SDS/PAGE. Note: To save a space, only a few fractions from the top of the gradients and the membrane pellet are shown. The assays were performed either by Coomassie blue staining (A), immunoblotting with SecA antisera (B, D–G), or autoradiography (C). In A, the standard molecular mass markers (in kDa) and GroEL as a marker are shown on the left and on the right, respectively. Band a on the right is the membrane protein sedimenting together with GroEL.
As a test of our hypothesis, we have performed an immunoblot analysis and found that the FtsY antisera does not recognize the band a (not shown), whereas the SecA antisera does. SDS gels in Fig. 3 show the content of SecA in a few fractions from the top of sucrose gradients and also in the membrane pellet after incubation without or with GroEL. As was found for the 100-kDa band a stained by Coomassie blue (Fig. 3A), in the presence of GroEL, a majority of SecA released from the membranes ran together with GroEL maximally in fraction 3 (Fig. 3B). In the absence of GroEL, soluble SecA was found at the top of the gradient (maximally in fractions 1 or 2). The GroEL preparation itself contains no detectable impurity of SecA (not shown). The use of IMVs carrying [35S]SecA, plasmid borne and metabolically labeled, yields a similar result (Fig. 3C). Furthermore, the intensity of the band of [35S]SecA running together with GroEL is certainly higher than that for [35S]SecA released from the membranes in the absence of GroEL. This suggests that GroEL is able not only to interact with SecA but also to stimulate dissociation of SecA from the membranes. An analogous phenomenon is also observed when IMVs carrying the SecA51(Ts) mutant is used. After incubation at 37°C (Fig. 3D) and especially at “heat shock” temperature, 42°C (Fig. 3E), where in the absence of GroEL, SecA51(Ts) does not dissociate at all, in the presence of GroEL a significant portion of the mutant appears to release from the membranes and sediment together with GroEL.
To further characterize an interaction of GroEL with SecA, some additional controls are presented. In particular, similar to the binding of GroEL to IMVs (Table 1), the interaction of GroEL with SecA51(Ts) is prevented by ATP (Fig. 3F) or EDTA (not shown). [Notice that the increase of the mutant at the top of the control gradient observed upon addition of ATP to IMVs (Fig. 3F, −GroEL) is consistent with observations of others (20, 22, 36) that ATP facilitates dissociation of SecA from the membrane.] Finally, incubation of GroEL with the membrane-depleted cellular extract does not reveal interaction of GroEL with cytoplasmic fraction of wt-SecA (Fig. 3G) or SecA51(Ts) (not shown). This is consistent with the findings of others (37) that GroEL has no affinity for the isolated SecA.
Thus, it can be concluded that GroEL interacts with the membrane-associated SecA and could “pull” it off the membranes. The “pulling” phenomenon is clearly seen for the wt-[35S]SecA, plasmid-borne and metabolically radiolabeled, or SecA51(Ts) mutant. Surprisingly, this effect is not observed for the unlabelled wt-SecA originated from chromosome (Fig. 3B). It seems clear that the GroEL-promoted release of this SecA a priori cannot be effective because a lot of SecA dissociates from the membrane regardless of GroEL (Fig. 3B, −GroEL). The observed distinction between the chromosomally produced SecA and the plasmid-borne [35S]SecA in their affinity for the membrane is unclear now and could be caused by non-identity of the membranes used. In particular, the IMVs and [35S]IMVs were isolated from different E. coli strains (MC4100 and T184, respectively), so that the conditions of growth of the cells from these strains differ considerably (Materials and Methods).
Cross-Linking of GroEL to SecA on the Membrane.
To confirm that GroEL indeed interacts with the membrane-embedded SecA, a cross-linking approach has been employed by using a cleavable, photoactivable, bifunctional reagent, TDPDP-ONSu (Fig. 4A and ref. 23). This cross-linker consists of (i) ONSu-activated carboxyl group, which allows incorporation of a TDPDP group into the protein via its specific interaction with amino groups; (ii) aryldiazirine group TDPDP with λmax ≈ 350 nm, mild UV irradiation of which generates a highly reactive carbene radical assumed to attack any chemical bond in the nearest neighborhood; and (iii) cis-diol group, which connects the photoactivable group with the activated carboxyl and is easily cleaved by periodate oxidation.
Cross-linking reaction is performed in two steps. In the first step, TDPDP groups are coupled to GroEL followed by removal of the excess reagent. In the second step, the photoactivable derivative of GroEL is incubated at 37°C with the membranes, carrying wt-[35S]SecA originated from plasmid ([35S]IMVs) or wt-SecA originated from chromosome (IMVs), and irradiated on ice. In the case of [35S]IMVs, the light-induced cross-linking is detected by upward shifting of radioactive material on an SDS gel. As seen in Fig. 4B, only under conditions stimulating the binding of GroEL to IMVs (i.e., in the presence of Mg ions and not of EDTA or ATP plus GroES), irradiation of the GroEL/IMVs mixture yields a cross-linked product X. This indicates that the cross-linking is a consequence of targeting of GroEL to IMVs and not of accidental collision of them. As seen, a molecular mass of the cross-linked product exceeds considerably a sum of those of GroEL subunit (56 kDa) and SecA (102 kDa). Such effect apparently is caused by the GroEL particle harboring approximately four photoactivable groups per subunit (Materials and Methods), and irradiation yields an inter-subunit cross-linking within the GroEL particle simultaneously and additionally to the cross-linking of GroEL to SecA (not shown). To confirm that GroEL cross-links to SecA and not to some radioactive impurities seen below the band of SecA (Fig. 4B), the irradiated [35S]IMVs were sedimented and solubilized by a nonionic detergent DM. The GroEL carrying the cross-linked radioactive label was then isolated by centrifugation in a sucrose gradient containing the detergent and analyzed by SDS/PAGE before or after periodate treatment. As seen in Fig. 4C, the periodate cleavage of a cis-diol bond between the GroEL and its membrane partner moieties within the cross-linked product shows downward change in position of radioactive band on an SDS gel so that the band X (lane 1) quantitatively converts into a band running at a position corresponding to SecA (lane 2). This means that the membrane-embedded radioactive component cross-linked to GroEL is indeed the SecA.
In the case of IMVs containing wt-SecA originated from chromosome, GroEL-SecA cross-linking product X was detected by immunoblotting with SecA antisera. As shown in Fig. 4D the appearance of band X requires interaction of GroEL with IMVs (lane 3). This band is not seen in the absence of irradiation (lane 1) or at irradiation in the presence of ATP plus GroES, which, as shown above, inhibit the GroEL-IMVs interaction (lane 5). The periodate treatment cleaves cross-linking product X and yields the SecA, which is detected by increasing intensity of the band SecA (lane 4).
Taken together, all of these observations are legitimate grounds to conclude that GroEL can bind to the membrane via the membrane-bound SecA, although participation of other unidentified membrane components is not excluded.
DISCUSSION
This study demonstrates the binding of GroEL to cytoplasmic membrane of E. coli and reveals participation of the membrane-associated protein SecA in this interaction. In some cases, after coming into contact with SecA, GroEL could stimulate its release from the membrane. It should be emphasized that all features, observed here and characterizing the targeting and interaction of GroEL with SecA on the membrane, correlate with the following well known properties of SecA:
(i) SecA is a membrane protein facing to cytoplasm (27). The conformation of SecA bound to the membrane is remarkably unfolded compared with that in solution (20, 36, 38, 39). This explains why GroEL prefers to interact with the membrane-bound and not with the soluble SecA (Fig. 3).
(ii) SecA is thermally denatured with a midpoint close to 42°C (39) that could cause a jump in binding of GroEL to the membrane (Fig. 2). Moreover, thermally induced denaturation of SecA is irreversible (40), which could be reflected in an inability of IMVs, preheated at 45°C, to bind GroEL at lower temperature (30°C, see Fig. 1C).
(iii) SecA exists and functions as a homodimer (34, 41, 42). This property of SecA points to its potential opportunity to interact with GroEL with positive cooperativity (Fig. 2).
(iv) Defects in protein secretion caused by mutation in SecA, in particular, by SecA51(Ts) mutant, are suppressed by overexpression of GroEL (11–13), suggesting that suppression is caused by interaction of GroEL with SecA.
(v) SecA is a very dynamic protein, because cytoplasmic, peripheral, and integral membrane forms of it have been detected (33, 34). This property of SecA correlates with an ability of GroEL to participate in partition of SecA between the membrane and cytoplasm. Furthermore, unlike wt-SecA (34, 43), the SecA51(Ts) mutant is shown to possess an inherent high affinity for the membrane, accumulating there (44). Therefore, a weakening the SecA51(Ts)-membrane association by GroEL and, by this, an adjusting of the membrane affinity for the mutant to that for wt-SecA seems a likely mechanism by which GroEL repairs the functional defect in SecA51(Ts) (11–13). Taken together, our data are in favor of the model according to which a dynamic membrane binding/release cycling of SecA is essential for its biological function (20, 36, 44, 45).
(vi) GroEL interacts not only with loosely membrane-bound SecA (Fig. 3), but also with the SecA embedded deeply into the membrane. In particular, the GroEL-SecA cross-linking products were isolated by solubilization of the membrane by DM (Fig. 4).
It is obvious that our procedures, indicating an interaction of GroEL with the membrane-bound SecA, do not allow us to conclude that SecA is the sole membrane component recognized by GroEL. Other not-yet-identified membrane proteins and also lipids could be involved in the membrane interaction with GroEL in addition to and independent of SecA. But a basic role of lipids in this process seems unlikely because some essential features of interaction of GroEL with cytoplasmic membranes of E. coli shown here (first, the effects of ATP and GroES) are contrary to those described recently for interaction of GroEL with model lipid mono- and bilayers (26).
Interaction of GroEL with the membrane-associated SecA might be physiologically relevant. One of the interacting partners, GroEL, has been shown to interact with newly synthesized secretory or membrane proteins and stimulate translocation across or insertion into the membrane (4–10). Another partner, SecA, is known to play a central role in translocation of a variety of secretory proteins across cytoplasmic membrane of E. coli by participation in formation and functioning of the protein-conducting channel (14, 46). Taken together with an ability of GroEL to interact with the membrane-bound SecA shown here, we speculate that GroEL as a chaperone might participate in functioning of the protein-conducting membrane apparatus by maintaining SecA in a functionally active conformation under a variety of conditions. In addition, GroEL also is able to modulate a cycle of SecA association with the membrane by stimulating release of SecA from the membrane. Finally, it is exciting to think that GroEL might act as a carrier and bring its passenger (secretory or membrane proteins) directly to the protein-conducting membrane channel. Our preliminary observations in vitro, that GroEL carrying the newly synthesized secretory protein proOmpA or polytopic membrane protein lactose permease can interact with IMVs followed by the ATP-promoted delivering of the bound protein to the membrane, are in favor of the carrier function of GroEL. Future experiments must examine the plausibility of our proposals and describe the mechanism by which GroEL participates in the membrane targeting of secretory and/or integral membrane proteins.
Acknowledgments
We are grateful to Don Oliver for generous gifts of SecA antisera and pT7SecA2, Eitan Bibi for E. coli strains MM52 and T184 and FtsY antisera, and Dmitri Bochkariov and Alexander Kogon for cross-linker TDPDP. We thank Ron Kaback, Steve Karlish, and Eitan Bibi for valuable discussions and critical reading of the manuscript. This research was supported by MINERVA Foundation, Germany.
ABBREVIATIONS
- IMVs
inverted cytoplasmic membrane vesicles
- TEA
triethanolamine
- TDPDP-ONSu
3-{3-[3-(trifluoromethyl)diazirin-3-yl]phenyl}-2,3-dihydroxypropionic acid N-hydroxysuccinimide ester
- DM
dodecylmaltoside
- wt
wild type
References
- 1.Buchner J. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 2.Lorimer G H. FASEB J. 1996;10:5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- 3.Fenton W A, Horwich A L. Protein Sci. 1997;6:743–760. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkareva E S, Lissin N M, Girshovich A S. Nature (London) 1988;336:254–257. doi: 10.1038/336254a0. [DOI] [PubMed] [Google Scholar]
- 5.Lecker S, Lill R, Ziegelhoffer T, Georgopoulos C, Bassford P J, Kumamoto C A, Wickner W. EMBO J. 1989;8:2703–2709. doi: 10.1002/j.1460-2075.1989.tb08411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battistoni A, Mazzetti A P, Petruzzelli R, Muramatsu M, Federici G, Ricci G, Lo Bello M. Protein Exp Purif. 1995;6:579–587. doi: 10.1006/prep.1995.1076. [DOI] [PubMed] [Google Scholar]
- 7.Shirai Y, Akiyama Y, Ito K. J Bacteriol. 1996;178:1141–1145. doi: 10.1128/jb.178.4.1141-1145.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkareva E S, Seluanov A, Bibi E, Girshovich A S. J Biol Chem. 1996;271:22256–22261. doi: 10.1074/jbc.271.36.22256. [DOI] [PubMed] [Google Scholar]
- 9.Meryandini A, Drews G. Photosynth Res. 1996;47:21–31. doi: 10.1007/BF00017750. [DOI] [PubMed] [Google Scholar]
- 10.Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips G J, Silhavy T J. Nature (London) 1990;344:882–884. doi: 10.1038/344882a0. [DOI] [PubMed] [Google Scholar]
- 12.Van Dyk T K, Gatenby A A, LaRossa R A. Nature (London) 1989;342:451–453. doi: 10.1038/342451a0. [DOI] [PubMed] [Google Scholar]
- 13.Danese P N, Murphy C K, Silhavy T J. J Bacteriol. 1995;177:4969–4973. doi: 10.1128/jb.177.17.4969-4973.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueguchi C, Ito K. J Bacteriol. 1992;174:1454–1461. doi: 10.1128/jb.174.5.1454-1461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 16.Teather R M, Bramhall J, Riede I, Wright J K, Furst M, Aichele G, Wilhelm U, Overath P. Eur J Biochem. 1980;108:223–231. doi: 10.1111/j.1432-1033.1980.tb04715.x. [DOI] [PubMed] [Google Scholar]
- 17.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell C, Oliver D. Mol Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 19.Bochkareva E S, Lissin N M, Flynn G C, Rothman J E, Girshovich A S. J Biol Chem. 1992;267:6796–6800. [PubMed] [Google Scholar]
- 20.Rajapandi T, Oliver D. Mol Microbiol. 1996;20:43–51. doi: 10.1111/j.1365-2958.1996.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 21.Tai P C, Tian G, Xu H, Lian J P, Yu J N. Methods Cell Biol. 1991;34:167–187. [PubMed] [Google Scholar]
- 22.Chen X, Xu H, Tai P C. J Biol Chem. 1996;271:29698–29706. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 23.Bochkariov D E, Kogon A A. Anal Biochem. 1992;204:90–95. doi: 10.1016/0003-2697(92)90144-v. [DOI] [PubMed] [Google Scholar]
- 24.Means G E, Feeney R E. Chemical Modification of Proteins. Inc., San Francisco: Holden–Day; 1971. p. 217. [Google Scholar]
- 25.Khandekar S S, Bettencourt B M, Kelley K C, Recny M A. Protein Exp Purif. 1993;4:580–584. doi: 10.1006/prep.1993.1076. [DOI] [PubMed] [Google Scholar]
- 26.Torok Z, Horvath I, Goloubinoff P, Kovacs E, Glatz A, Balogh G, Vigh L. Proc Natl Acad Sci USA. 1997;94:2192–2197. doi: 10.1073/pnas.94.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver D B, Beckwith J. Cell. 1982;30:311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- 28.Meyenburg K, Jorgensen B B, Deurs B. EMBO J. 1984;3:1791–1797. [Google Scholar]
- 29.Matsuyama S-i, Fujita Y, Sagara K, Mizushima S. Biochim Biophys Acta. 1992;1122:77–84. doi: 10.1016/0167-4838(92)90130-6. [DOI] [PubMed] [Google Scholar]
- 30.Melchior D L. Curr Top Membr Transp. 1982;17:263–316. [Google Scholar]
- 31.Nakayama H, Mitsui T, Nishihara M, Kito M. Biochim Biophys Acta. 1980;601:1–10. doi: 10.1016/0005-2736(80)90508-8. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt M G, Rollo E E, Grodberg J, Oliver D B. J Bacteriol. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebke H H. J Bacteriol. 1987;169:1174–1181. doi: 10.1128/jb.169.3.1174-1181.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabelli R J, Dolan K M, Qian L, Oliver D B. J Biol Chem. 1991;266:24421–24427. [PubMed] [Google Scholar]
- 35.Luirink J, Hagen-Jongman C M, der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Economou A, Wickner W. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 37.Hartl F U, Lecker S, Schiebel E, Hendrick J P, Wickner W. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 38.Shinkai A, Mei L H, Tokuda H, Mizushima S. J Biol Chem. 1991;266:5827–5833. [PubMed] [Google Scholar]
- 39.Ulbrandt N D, London E, Oliver D B. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 40.den Blaauwen T, Fekkes P, de Wit J G, Kuiper W, Driessen A J M. Biochemistry. 1996;35:11994–12004. doi: 10.1021/bi9605088. [DOI] [PubMed] [Google Scholar]
- 41.Akita M, Shinkai A, Matsuyama S-i, Mizushima S. Biochem Biophys Res Commun. 1991;174:211–216. doi: 10.1016/0006-291x(91)90507-4. [DOI] [PubMed] [Google Scholar]
- 42.Driessen A J M. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 43.Chun S-Y, Randall L L. J Bacteriol. 1994;176:4197–4203. doi: 10.1128/jb.176.14.4197-4203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver D B, Cabelli R J, Jarosik G P. J Bioenerg Biomembr. 1990;22:311–336. doi: 10.1007/BF00763170. [DOI] [PubMed] [Google Scholar]
- 45.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 46.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]