Abstract
Background
CatSper1 and CatSper2 are two recently identified channel-like proteins, which show sperm specific expression patterns. Through targeted mutagenesis in the mouse, CatSper1 has been shown to be required for fertility, sperm motility and for cAMP induced Ca2+ current in sperm. Both channels resemble a single pore forming repeat from a four repeat voltage dependent Ca2+ /Na+ channel. However, neither CatSper1 or CatSper2 have been shown to function as cation channels when transfected into cells, singly or in conjunction. As the pore forming units of voltage gated cation channels form a tetramer it has been suggested that the known CatSper proteins require additional subunits and/or interaction partners to function.
Results
Using in silico gene identification and prediction techniques, we have identified two further members of the CatSper family, CatSper3 and Catsper4. Each carries a single channel-forming domain with the predicted pore-loop containing the consensus sequence T×D×W. Each of the new CatSper genes has evidence for expression in the testis. Furthermore we identified coiled-coil protein-protein interaction domains in the C-terminal tails of each of the CatSper channels, implying that CatSper channels 1,2,3 and 4 may interact directly or indirectly to form a functional tetramer.
Conclusions
The topological and sequence relationship of CatSper1 and CatSper2 to the four repeat Ca2+ /Na+ channels suggested other members of this family may exist. We have identified a further two novel CatSper genes, conserved in both the human and mouse genomes. Furthermore, all four of the CatSper proteins are predicted to contain a common coiled-coil protein-protein interaction domain in their C-terminal tail. Coupled with expression data this leads to the hypothesis that the CatSper proteins form a functional hetero-tetrameric channel in sperm.
Background
Channel activities, particularly those of calcium channels, have been linked to the process of sperm maturation, motility and to the sperm egg interaction. A number of candidate channels have been associated with these processes, these include the voltage operated Ca2+ channels alpha1A, 1C and 1E plus beta subunits [1,2], the T-type voltage operated Ca2+ channels alpha 1G and 1H, cyclic nucleotide gated (CNG) channels [3] and the transient receptor potential (TRP) channel TRP2 [4], The evidence being primarily based on transcript and immuno-staining studies. However, none of the channels expressed in testes and sperm have been directly associated with sperm mobility. Recently, two novel channel-like proteins, CatSper1 and CatSper2 (Cation channel of Sperm), were identified to be specifically expressed in spermatozoa and to be linked to sperm mobility [[5,6]; reviewed in [7]].
The human and mouse CatSper channels, CatSper1 and CatSper2, both carry a single six-transmembrane spanning unit analogous to one of the four repeats found in voltage-dependent Ca2+ channels [5,6]. Analysis of the pore forming region within the repeat suggested that CatSper1 and 2 are Ca2+ selective [5,6]. Further evidence supporting channel activity has been provided for CatSper1 by gene-targeting experiments in the mouse [5]. Notably in sperm from the mice carrying two null alleles for the CatSper1 gene, cAMP and cGMP induced Ca2+ influx is lost. Moreover, CatSper1 has been shown to be required for normal sperm motility and egg penetration. However, attempts to define channel activity for CatSper1 and CatSper2 – singly or in conjunction – in heterologous expression systems have failed [5,6]. This suggests that additional factor or factors are required to form a functional channel. The fact that CatSper1 and 2 share features of a single repeat of a four repeat channel suggests that an additional two members might exist.
Here we describe the prediction of two additional CatSper channels in the human and mouse genomes, CatSper3 and CatSper4. Both channels contain a single six-transmembrane repeat domain, which contain the T×D×W pore-lining consensus sequence present in CatSper1 and CatSper2. Based on accompanying EST, cDNA library source information and Taqman data, both genes are expressed in testis. Furthermore, we noted that each CatSper channel is predicted to contain a coiled-coil motif, a protein-protein interaction interface, in its intra-cellular C-terminal tail. Based on a common expression pattern and the fact that each CatSper protein is predicted to contain a coiled-coil domain, we hypothesise that the CatSpers come together to form functional tetrameric channels either by direct interaction of their coiled-coil motif or through interaction with additional factors.
Methods
Identification of CatSper3 and 4
PSI-BLAST profiles were constructed from sequence alignments of the ion-transport domain of CatSper protein sequences and calcium channel protein sequences. These profiles were used as input to the PSI-BLAST algorithm [8] to search large human and mouse genome based protein databases for potential novel members of the CatSper protein family. Two GENSCAN [9] human gene predictions partially covering the ion transport domain region were identified. The GENEWISE program [10] was used to improve and extend the original gene predictions into full-length proteins using CatSper2 protein sequence as a template. Orthologous mouse CatSper3 and mouse CatSper4 sequences were identified by mapping syntenic regions of CatSper3 and CatSper4 human loci to the mouse genome. The GENEWISE program was again used to craft the final mouse proteins from their DNA, seeded by human CatSper3 and human CatSper4 predictions. Overlapping EST sequences listed in Table 5 were also used to improve predictions.
Table 5.
Representation of CATSPER 3 and 4 in sequence databases and associated tissue sources.
| Gene | Genomic | cDNA | EST | Patent |
| Human CATSPER3 | AC004764 | Not present | BI827754 901 bp (Brain-medulla) | AX358304 – WO0194412 (No Tissue distribution) |
| AI219834 337 bp (Pooled Testis-lung-B cell) | AX047619 – WO0066735 (Predominantly Testis by Northern blot) | |||
| AI027609 362 bp (Testis) | ||||
| AW003058 306 bp (Germ cell tumor) | ||||
| AW972257 417 bp (Colon cancer) | ||||
| AW593391 306 bp (Germ cell tumor) | ||||
| AW590264 306 bp (Germ cell tumor) | ||||
| AW003002 306 bp (Germ cell tumor) | ||||
| AW008956 258 bp (Colon) | ||||
| AW007549 256 bp (Colon) | ||||
| AA527520 255 bp (Colon) | ||||
| BX280235 212 bp (Pooled Testis-lung-B-cell) | ||||
| Mouse CATSPER3 | AC129780 | AK014942 1312 bp (Adult testis) | BY714458 951 bp (Adult testis) | Not present |
| BB679726 390 bp (Adult testis) | ||||
| BY510167 408 bp (Bone marrow macrophage) | ||||
| CA465993 758 bp (Testis) | ||||
| BF147131 335 bp (Testis) | ||||
| AV043837 252 bp (Testis) | ||||
| AV280964 263 bp (Adult testis) | ||||
| BB017226 225 bp (Adult testis) | ||||
| BB013542 244 bp (Adult testis) | ||||
| Human CATSPER4 | AL355877 | Not present | AA421134 518 bp (Testis) | Not present |
| Mouse CATSPER4 | AL627314 | AK077145 1713 bp (Adult testis) | AV280157 622 bp (Adult testis) | Not present |
| BB617038 605 bp (Adult testis) | ||||
| BY096088 378 bp (Adult testis) | ||||
| BY088873 365 bp (Pooled adult tissue) | ||||
| BY454059 444 bp (Pooled adult tissue) | ||||
| BU961662 795 bp (Testis) | ||||
| BY459016 398 bp (Adult testis) | ||||
| AV269930 241 bp (Adult testis) | ||||
| AV281460 247 bp (Adult testis) | ||||
| AV263155 176 bp (Adult testis) |
Expression Analysis
Human RNA prepared from non-diseased organs was purchased from either Ambion Europe or Clontech. cDNA was prepared from 500 ng RNA using random hexamers and Multiscribe (Applied Biosystems), following manufacturer's instructions.
Oligonucleotide primers and probes were designed using Primer Express software (Applied Biosystems) with a GC-content of 40–60%, no G-nucleotide at the 5'-end of the probe, and no more than 4 contiguous Gs. Each primer and probe was analysed using BLAST (Basic Local Alignment Search Tool) [8]. Results confirmed that each oligonucleotide recognises the target sequence with a specificity >3 bp when compared to other known cDNA's or genomic sequence represented in the NCBI publicly available databases [11].
The sequence of the primers and probes directed against human CatSper4 exon 9 are as follows:
Forward primer: 5'-AAGGACATCCGCCAGATGTC-3'
Reverse primer: 5'-GGCACACCTTTTCCATGCTAA-3'
Probe: 5'-CAACAGCAAGACTTGCTCAGTGCGCT-3'
Expected amplicon size is 70 bp, a test PCR reaction was carried out under the following conditions; 40 cycles, 95°C 30 seconds, 58°C 30 seconds, 72°C 30 seconds. Expected amplicon size was confirmed on an agarose gel.
18S rRNA pre-optimised primers and probe were purchased from Applied Biosystems, Foster City, CA.
25 μl PCR reactions were carried out using TaqMan Universal Master Mix (Applied Biosystems) following manufacturer's instructions and as described in Lobenhofer et al [12].
Each sample reaction contained 100 nM Taqman probe; 300 nM forward primer; 900 nM reverse primer and 15 ng of cDNA template. Within each experiment, a standard curve was carried out of a typical tissue sample, from 50 ng to 0.78 ng of cDNA template. From this standard curve, the amount of actual starting target or 18S cDNA in each test sample was determined. The levels of target cDNA in each sample were normalised to the level of expression of target in a comparative sample. The levels of 18S cDNA in each sample were normalised to the level of expression of 18S in a comparative sample. The data was then represented as fold expression of target sequence normalised to 18S expression relative to the level of expression in the comparative sample, which was set arbitrarily to 1.
Characterisation of the CatSper channel family
CatSper3 and CatSper4 transmembrane regions were predicted using the TMHMM program [13] and delineated by analogy with other CatSper and calcium-channel family members. Coiled-coils were predicted using the COILS algorithm for each member of the CatSper channel family [14].
Sequence Alignments and Phylogenetic Tree
Sequence alignments of the CatSper protein family, and calcium channels were created using CLUSTALW multiple sequence alignment program [15] and hand-crafted using the JALVIEW sequence alignment editor [16]. Pairwise sequence identities presented in tables 6 and 7 were calculated using the pairwise sequence alignment algorithm present in the JALVIEW software.
Table 6.
Sequence identities shown between human CatSper family members
| Hsper1 | Hsper2 | Hsper3 | Hsper4 | |
| Hsper 1 | 100 | 21.95 | 22.18 | 27.08 |
| Hsper 2 | 21.95 | 100 | 23.33 | 26.42 |
| Hsper 3 | 22.18 | 23.33 | 100 | 23.93 |
| Hsper 4 | 27.08 | 26.42 | 23.93 | 100 |
Table 7.
Sequence identities shown between calcium channel repeats within the same gene and orthologous repeats. CcaH and CcaG sequences correspond with Swissprot identifiers CCAH_HUMAN and CCAG_HUMAN T type calcium channels. CcaC and CcaS represent Swissprot sequences CCAS_HUMAN and CCAC_HUMAN L type calcium channels.
| CcaH I | CcaH II | CcaH III | CcaH IV | CcaG I | CcaG II | CcaG III | CcaG IV | |
| CcaH I | 100 | 23.24 | 27.87 | 20.69 | 75.68 | 22.41 | 27.79 | 23.26 |
| CcaH II | 23.24 | 100 | 26.10 | 26.77 | 22.41 | 88.41 | 27.94 | 26.29 |
| CcaH III | 27.87 | 26.10 | 100 | 28.36 | 27.79 | 27.94 | 80.15 | 29.45 |
| CcaH IV | 20.69 | 26.77 | 28.36 | 100 | 23.26 | 26.29 | 29.45 | 79.92 |
| CcaS I | CcaS II | CcaS III | CcaS IV | CcaC I | CcaC II | CcaC III | CcaC IV | |
| CcaS I | 100 | 23.26 | 28.71 | 22.62 | 71.96 | 25.08 | 24.38 | 19.94 |
| CcaS II | 23.26 | 100 | 25.98 | 26.28 | 25.08 | 77.08 | 23.33 | 24.32 |
| CcaS III | 28.71 | 25.98 | 100 | 21.24 | 24.38 | 23.33 | 70.71 | 22.11 |
| CcaS IV | 22.26 | 26.28 | 21.24 | 100 | 19.94 | 24.32 | 22.11 | 67.91 |
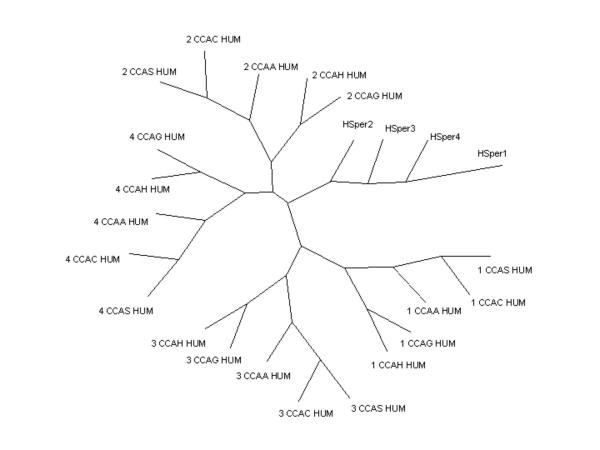
The phylogenetic tree shown in figure 8 was constructed from sequence alignments of the CatSper protein family with calcium channel sequences CCAA_HUMAN, CCAH_HUMAN, CCAG_HUMAN, CCAS_HUMAN, CCAC_HUMAN over the ion-transport domain region. PHYLIP [16] PROT-PARS maximum parsimony programme was used to build 1000 bootstrap trees from the sequence alignment. The final tree was obtained using the CONSENSE programme to select the best tree by majority rule.
Figure 8.
Un-rooted phylogenetic tree showing CatSper family members with selected T- and L-type calcium channels. Repeats 1–4 of selected human L- and T-type calcium channels; CCAA_Human – calcium channel alpha1A (SWISSPROT O00555), CCAC_Human – calcium channel alpha1C (SWISSPROT Q13936), CCAG_Human – calcium channel alpha1G (SWISSPROT O43497), CCAH_Human – calcium channel 1H (SWISSPROT O95180) and CCAS_Human – calcium channel alpha1S (SWISSPROT Q13698).
Results
Identification of CatSper 3 and 4
As part of an on going program to identify novel ion channel encoding genes, ion-channel family sequence profiles have been used to search sets of human gene predictions. Two initial GENSCAN [9] predictions of 264 aa and 185 aa mapping to human chromosomes 5q31.1 and 1p35.3 respectively contain features related to the previously described CatSper genes – namely a single ion-transport domain and a pore-loop containing the consensus T × D × W. The predictions have been hand polished using a combination of GENSCAN and GENEWISE [10] analysis, coupled with Expressed Sequence Tag (EST) data and homology between human and mouse chromosomes to obtain full-length gene models.
Human CatSper3 has eight coding exons, spanning a region of 43.7 kb giving rise to an open reading frame of 398 aa (Figure 1 and Table 1). The human Catsper3 prediction is supported by 12 ESTs from mixed tissue types include germ cell tumors and testis (Table 5). Human Catsper3 also appears in the patent literature, Lexicon Genetics: WO200066735 and Millenium Pharamaceuticals: WO200194412; as a novel human ion-channel cloned from a testis library – Lexicon Genetics: WO200066735; and as a putative sodium channel – Millenium Pharamaceuticals: WO200194412 (Table 5). Available tissue distribution information from ESTs and the patent literature show that human CatSper3 is predominantly a testis derived transcript although there is also a suggestion that transcripts are found in other tissues (Table 5).
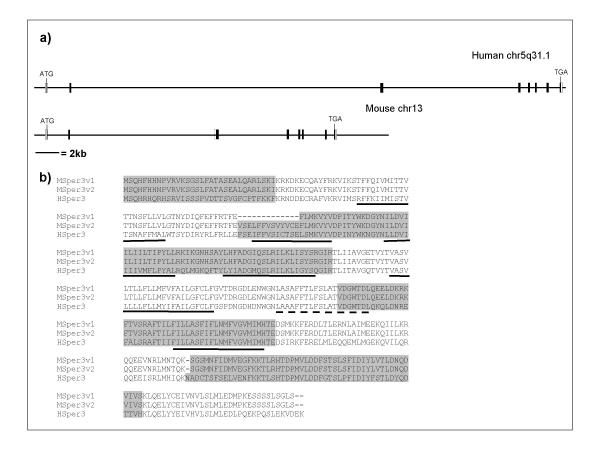
Figure 1.
Genomic organisation of the human and mouse CatSper3 genes. (a) Schematic of human and mouse CatSper3 genes on human chromosome 5q31.1 and mouse chromosome 13 respectively. Horizontal line represent human genome assembly NCBI 31 and mouse genome assembly NCBI 03, filled boxes represent coding regions, un-filled boxes represent non-coding regions (b) Comparison of exon boundaries between human and mouse genes, exons are shaded alternately, MSper3v1 and MSper3v2, represent the predicted splice variants of mouse CatSper3. Predicted transmembrane regions are underlined, the pore forming region is underlined with a dashed line.
Table 1.
Exon/intron boundaries of human CATSPER3
| Exon | Exon Length bp | Splice Acceptor* | Splice Donor* | Intron Length bp |
| 1 | 167 | Not Applicable | AATTTAAgtaaata | 1848 |
| 2 | 154 | ttgacagGAGGAAC | TCTTGAGgtaagca | 26179 |
| 3 | 240 | cctgtagTTCTCGG | CATCCGGgtgagtg | 11443 |
| 4 | 183 | cttgcagACGCTGA | GGCCACGgtactgt | 700 |
| 5 | 141 | tgagcagGTTGATG | CACAGAGgtgaggc | 388 |
| 6 | 120 | tctctagGACTCCA | CATACAGgtgagtg | 881 |
| 7 | 158 | tctctagAAAAATG | TCCACAAgtcagtt | 989 |
| 8 | >176 | ttttcagGCTTCAA | Not Applicable | N/A |
* Intronic sequences are in lower case
In comparison to human CatSper3, the mouse CatSper3 gene spans a region of approximately 24 kb on mouse chromosome 13 – however, gaps remain in the current mouse genome assembly and therefore intron sizes can not be determined precisely (Figure 1 and Table 2). Based on GENEWISE comparison of the mouse genomic sequence with the human CatSper3 ORF, the mouse CatSper3 gene is predicted to encode an open reading frame of 395 aa. Mouse CatSper 3 is also represented by a RIKEN cDNA clone (AK014942) [18] from an adult mouse testis library; however, this encodes a shorter protein of 382 aa. This is due to use of an alternative splice acceptor site within the third exon (Msper3v1:Figure 1b and Table 2). This shorter version is predicted to have a truncated 2nd transmembrane helices and, therefore, is unlikely to form a functional ion channel.
Table 2.
Exon/intron boundaries of mouse CATSPER3
| Exon | Exon Length bp | Splice Acceptor* | Splice Donor* | Intron Length bp |
| 1 | 181 | Not Applicable | AGATTAAgtaagta | 1737# |
| 2 | 154 | ttggcagGAGGAAG | CTTTGAGgtgagct | 12363v1/12324v2# |
| 3v1 | 201 | ctgcgagTTCCTCA | CATCAGGgtgagtc | 5803 |
| 3v2 | 240 | actgcagGTCTCAG | CATCAGGgtgagtc | 5803 |
| 4 | 183 | cttgcagACACTCA | GGCCACGgtactga | 746 |
| 5 | 141 | tgaccagGTTGATG | CACGGAGgtgagga | 381 |
| 6 | 120 | gtgttagGATTCCA | CACACAGgtcggat | 1662 |
| 7 | 155 | cctctagAAAAGTG | TCAGCAAgtgagtt | 647 |
| 8 | >175 | gttctagGCTTCAG | Not Applicable | N/A |
* Intronic sequences are in lower case. # Estimated intron size due to gaps in genome assembly.
Notably, exon 1 of human CatSper3 lies within the 3'UTR of the DCOHM gene (dimerisation cofactor of hepatocyte nuclear factor from muscle: Genbank AF499009)[19] such that the two genes are in a head-to-tail orientation. The DCOHM cDNA has been isolated from muscle and kidney libraries, whereas available tissue distribution information for human CatSper3 points to a predominantly testis specific expression. Therefore transcriptional interference is unlikely to occur between the two genes. Using the human DCOHM as a query sequence, an ORF of 90% sequence identity can also be found in the mouse genome 8.5 kb upstream of the mouse CatSper3 start codon. Therefore a similar gene arrangement to the human loci exists in the mouse (data not shown). However, we do have any information relating to the extent of the mouse DCOHM 3'UTR as this has not yet been cloned. Therefore, the mouse DCOHM gene may or may not extend over the mouse CatSper3 coding exons.
The human CatSper4 gene is predicted to span a region of 12 kb and be comprised of 10 coding exons, (Figure 2a and Table 3). Human CatSper4 is only partially represented by a single EST originating from a testis library. In contrast mouse CatSper4 is present in the databases as a RIKEN testis derived cDNA (AK077145) [18] and is also represented by ten ESTs, all of which are either testis derived or derived from a pooled library containing testis material.
Figure 2.
Genomic organisation of the human and mouse CatSper4 genes. (a) Gene structure of human and mouse CatSper4 genes on human chromosome 1p35.3 and mouse chromosome 4 band D3 respectively. Horizontal line represent human genome assembly NCBI 31 and mouse genome assembly NCBI 03, filled boxes represent coding regions, un-filled boxes represent non-coding regions. (b) Comparison of exon boundaries between human and mouse genes, exons are shaded alternately. Predicted transmembrane regions are underlined, the pore forming region is underlined with a dashed line.
Table 3.
Exon/intron boundaries of human CATSPER4
| Exon | Exon Length bp | Splice Acceptor* | Splice Donor* | Intron Length bp |
| 1 | >213 | Not Applicable | CAAAGCGgtaagga | 445 |
| 2 | 144 | cttccagGACGCCT | GGACCAGgtgggat | 2355 |
| 3 | 102 | catgtagAAACACT | CTGGAAGgtgagat | 3796 |
| 4 | 98 | tccccagGACGGCT | CTCTCAGgtgagcg | 172 |
| 5 | 121 | gccacagGGCGCTT | CATGCTGgtcagtg | 207 |
| 6 | 134 | ctgacagGTTTTTT | ACTTCCAgtgagtg | 1463 |
| 7 | 175 | tctgcagGACAGAG | TAGTGAGgtgcgtg | 770 |
| 8 | 212 | ttcccagACAGGCG | TCAACATgtagggg | 311 |
| 9 | 166 | cccccagGATTGTT | GGAAAAGgtgtgcc | 968 |
| 10 | >54 | tttgcagGTTCATG | Not Applicable | N/A |
* Intronic sequences are in lower case.
The mouse CatSper4 gene is located on mouse chromosome 4 band D3, it spans a region of 15.3 kb. Unlike the human CatSper4 the mouse gene possesses 11 coding exons, the gene structure varying with respect to the human in the first two exons (Figure 2 and Table 4) otherwise exon/intron boundaries are conserved Figure 2b.
Table 4.
Exon/intron boundaries of mouse CATSPER4
| Exon | Exon Length bp | Splice Acceptor* | Splice Donor* | Intron Length bp |
| 1 | >111 | Not Applicable | TCCTAGGgtgggtg | 43 |
| 2 | 75 | tctgcagAGAAAAG | GAAGAAGgtaagga | 401 |
| 3 | 144 | cttccagGATGCCT | CGGTCAGgtgggat | 4939 |
| 4 | 102 | catgtagAAACATT | CTGGAAGgtgagct | 3012 |
| 5 | 98 | tattcagGATGGCT | CTCTCAGgtgagtt | 146 |
| 6 | 121 | cccacagGGTGCTC | CATGCTGgtgagtg | 200 |
| 7 | 134 | ctgacagGTATTCT | ACTTCCAgtgagta | 2486 |
| 8 | 175 | ttcccagGATGGAT | TACTGAGgtgagcc | 1272 |
| 9 | 209 | ttcccagACAGAAG | TCAACATgtagggg | 300 |
| 10 | 154 | cccccagGATCGTG | GGAAAAGgtgcgtg | 673 |
| 11 | 400(incl 3'UTR) | tttgcagGTGTCTG | Not Applicable | N/A |
* Intronic sequences are in lower case. # Estimated intron size due to gaps in genome assembly
Having found two further members of the CatSper family in human and mouse genomes a search for orthologues in Fugu rubripes and Danio rerio was carried out. Searching with CatSper sequences against raw genomic sequence (TBLASTN) [8] and ENSEMBL[20] protein predictions (BLASTP)[8] failed to identify any orthologues. Furthermore we failed to identify any channel-like sequence of less than 400 aa containing a pore-forming region of the consensus T×D×W. Given the current coverage of the Fugu genome it is surprising that no CatSper-like sequences were identified.
Tissue distribution of Human CatSper4
The previously described CatSper sequences, CatSper1 and CatSper2 are expressed in testis and more specifically spermatocytes. Data from ESTs and patent literature suggest that CatSper3 also shares a common expression profile. As there was limited evidence supporting the human CatSper4 transcript we carried out a Taqman quantitative PCR analysis to address expression of the Human Catsper4 gene. A primer probe set was designed within exon 9 of human CatSper 4 sequence and tissue expression profiling was carried out in 18 normal human tissues as described in Materials and Methods. Figure 3a shows the correct amplicon size for primers directed against human CatSper4 exon9 and figure 3b shows the normalised level of expression of CatSper 4 in the 18 tissues. These data confirm the prediction of testis specific expression. Low expression levels were detected in placenta and lung, whereas no significant expression was detected in any other tissue.
Figure 3.
Normalised expression of Human CatSper4 in 18 normal human tissues. (a) Amplicon size of human CatSper4, amplified from exon9. Lane 1 no template control, Lane 2 15 ng Testis cDNA, Lane 3 40 ng testis cDNA, Lane 4 DNA size marker ascending in 100 bp intervals from 100 bp upwards (Eurogentec). (b) Levels of human CatSper4 mRNA in 18 normal human tissues were determined using Taqman quantitative RT-PCR. Each sample was quantitated in 3 individual experiments, the mean ± SEM for the multiple experiments are shown. Tissue names followed by (2) represents an alternative RNA supplier.
Protein features
CatSper 3 and 4 are predicted to contain 6 transmembrane regions denoted S1–S6 in figures 4a and 4b. S1–S4 are close together joined by short loop regions. A longer loop region separates S5 and S6 and contains a short conserved hydrophobic stretch – see Figure 4 for topology cartoon of the CatSper family. The arrangement of these transmembrane helices is characteristic of the voltage gated channel ion transport domain found in voltage-gated K+, Ca2+ and Na+ channels, and reported for the other members of the CatSper channel family [5,6]. This domain comprises 6 transmembrane helices with a hydrophobic channel pore loop and voltage-sensing region.
Figure 4.
Topology of the CatSper channels. (a) Topology diagram of CatSper1 protein based on Ren et al. (b) Topology diagram of CatSper2,3 and 4 proteins.
The voltage sensor lies within S4 transmembrane helix and is involved in channel activation via positively charged residues positioned every 3–4 amino acids [21]. Sequence alignment of S4 helices of selected voltage gated Ca2+ channels with CatSper family (Figure 6a) shows that a pattern of regular repeating basic residues (arginine/lysine) are also present in the CatSper1 and CatSper2 S4 helices. However, in CatSper3 and CatSper4 subunits the repeating charged residues are conserved to a lesser extent with only two of the four charged residues found, suggesting a reduced voltage dependent mechanism of activation [21].
Figure 6.
Alignment of Human CatSper voltage sensor and pore forming regions with selected L- and T-type calcium channels. (a) Voltage sensor region of CatSper human protein sequences aligned with repeats 1–4 of selected human L- and T-type calcium channels; CCAA_Human – calcium channel alpha1A (SWISSPROT O00555), CCAS_Human – calcium channel 1S (SWISSPROT Q13698) and CCAH_Human – calcium channel 1H (SWISSPROT O95180). (b) Pore selectivity region of CatSper ion channel family aligned with selected human L- and T-type calcium channels ion transport repeats.
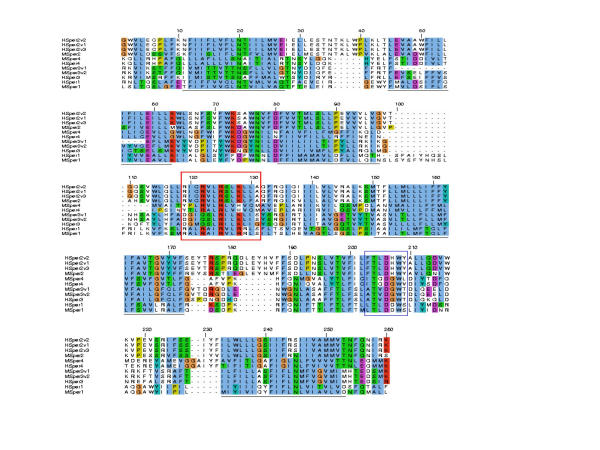
Ion specificity is determined by a pore consensus sequence [T/S] × [D/E] × W in voltage gated Calcium channels [22]. Sequence analysis of this region in the CatSpers highlights the presence of a similar conserved motif T×D×W (Figure 5 and 6b) suggesting that the CatSper ion channels may be selective for calcium ions, as previously discussed for CatSper1 [5]. BLASTP homology searches also link the CatSpers most closely with the T-type Calcium channels.
Figure 5.
Multiple sequence alignment of the CatSper ion channel family ion-transport domain. Transmembrane regions are underlined in black and the S4 voltage sensor transmembrane helix is highlighted in red. The channel pore consensus sequence motif is boxed in blue. Genbank accession codes for the published Catsper genes are as follows: MSper1 AF407332, HSper1 AF407333, MSper2 AF411816, HSper2v1 AF411817, HSper2v2 AF411818, HSper2v3 AF411819.
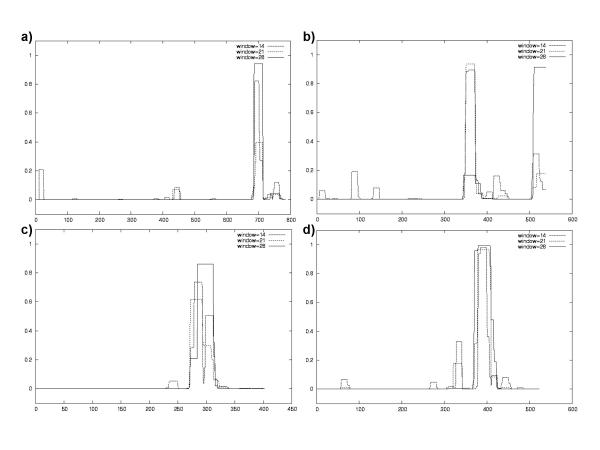
Each member of the CatSper family contains a coiled-coil domain at its C terminus as predicted by COILS programme [14] and shown in Figure 7. Coiled-coils are well characterised as potential protein-protein interaction domains. They have also been found in multi-pass membrane proteins such as GABABR1 and GABABR2 to be the site of receptor dimerisation [23,24]. Coiled-coils have also been found in multi-protein complexes such as the SNARE complex [25]. The identification of a common protein-protein interaction domain in all four of the CatSper proteins within the context of a common expression pattern and relationship to four-repeat calcium channels suggest that the CatSper ion channel subunits assemble as tetramers.
Figure 7.
Results from CatSper channel coiled-coil predictions. (a) CatSper1 human coiled-coil prediction, (b) CatSper2 human coiled-coil prediction, (c) CatSper3 coiled-coil prediction and (d) CatSper4 coiled-coil prediction. X-axis, amino acid residue numbering of query sequence. Y-axis, probability score for a sequence adopting a coiled-coil configuration calculated for a scanning window of 14, 21 or 28 amino acid residues.
CatSper3 and CatSper4 extend the Calcium channel family
The CatSper ion channel subunits are distant in sequence relationship; sequence identity ranges between 21.6% and 26.5% across the ion transport domain (Table 6). This low sequence identity is in contrast with that observed for the voltage-gated sodium and calcium channel families. Calcium L-type calcium channels generally share ~25% sequence identity over full their length sequence and upwards of 75% sequence identity between their corresponding ion-transport repeat regions (Table 7). These observations are further supported by the phylogenetic tree (Figure 8) which shows that each repeat is more closely related to its analogous repeat in a paralagoue than to the other repeats in the same gene unit, i.e. repeat I in alpha 1S is more closely related to repeat I in alpha 1T than to repeat II in alpha 1S. Figure 9 shows the repeat topology of a voltage-gated cation channel. In addition, repeats 1 and 3, and repeats 2 and 4 share common ancestry with all four repeats stemming from a single common ancestor (Figure 8). In contrast, the CatSper family members do not associate with any one particular repeat, this therefore raises questions over the detailed evolutionary history of the CatSper family.
Figure 9.
Topology diagram for the L-type and T-type four repeat voltage gated calcium channel families.
Discussion
Here, we applied bioinformatic tools in a focused approach to identify and characterise novel ion-channel genes in both human and mouse genomes. We identified two genes, CatSper3 and CatSper4, which extend the CatSper ion channel-like family to four members in human and mouse. As previously described for CatSper1 and 2 [5,6], CatSper3 and 4 contain a single ion transport domain comprised of 6 transmembrane spanning regions, where the fourth transmembrane region resembles a voltage sensor and a pore forming region lies between transmembrane regions 5 and 6. The pore contains the consensus sequence T×D×W indicative of a probable calcium selective channel. Available expression data suggest that CatSper3 and 4 are present in testis and may also be found in other tissues. To date, CatSper1 and 2 have not shown channel activity when expressed in heterologous systems alone or when co-expressed. One explanation is that additional factors are required for full function. The identification of two more CatSper like channels both of which show expression in testis and both of which resemble single pore forming repeats from a multi-repeat channel, may well provide the missing factors required for a functional CatSper channel to be formed.
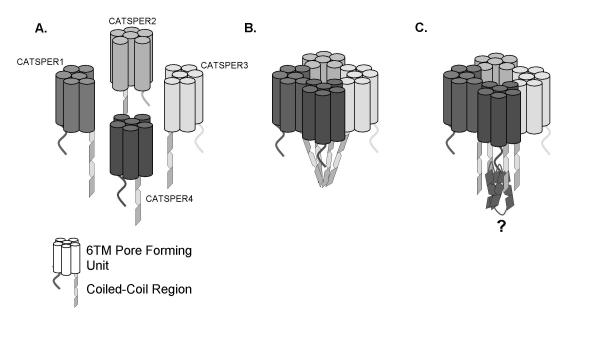
Additionally, through our bioinformatic analysis of the CatSper family we have annotated coiled-coil domains in all four of the CatSper channels. Alpha helical coiled-coil structural motifs are involved in subunit multimerisation of a large number of proteins. For example, the GABAB receptor assembly is mediated by short (~30 aa) parallel coiled-coil alpha helices in the C-terminal of the GABABR1 and GABABR2 receptors [24]. Coiled-coil domains can also mediate formation of large multi-protein complexes such as the SNARE complex whose core comprises a hetero-tetrameric coiled-coil [25]. Therefore a precedent exists for a four coiled-coil complex. Identification of the coiled-coil domains in the CatSper channels provides an experimentally testable mechanism for CatSper channel tetramerisation. This in theory could involve interaction at the coiled coil domain directly, or via intracellular accessory proteins that interact with the CatSper subunits via the coiled coil motif, anchoring the subunits together. A proposed model for subunit interaction is shown in Figure 10. One question raised by the CatSper1 knockout experiment is how are the channels regulated by cAMP/cGMP? We have searched for cyclic nucleotide binding sites on the CatSper subunits, however no likely domains have been identified, it is therefore possible that this property is conferred by an auxiliary subunit and therefore this would favour the model proposed in Figure 10c.
Figure 10.
Theoretical model for the formation of a CatSper hetero-terameric channel. (a) Diagramatic representation of CatSper subunits 1–4. (b) In this model a tetrameric channel is formed via direct interaction of the coiled-coil domains. (c) In this model the channel is brought together via an auxiliary protein or proteins, interaction being mediated via the coiled-coil domains.
The above model for CatSper subunits function and interactions could be tested in a variety of experiments. Function may be tested through targeted mutagenesis experiments of the new CatSper subunits in mice as described by Ren et al [5]. Expression of all four subunits in an heterologous expression system could be attempted with the aim of reconstituting a functional channel. To identifying interactions via the coiled-coil domain, the intracellular domain of the CatSper subunits could be used as the "bait" in the yest two-hybrid system. This system was successful in identifying the GABABR2 receptor as the co-receptor GABABR1 [23] via a coiled-coil domain. Certainly the identification of two further CatSper subunits provides further possibilities in which to test this family of protein's function in sperm mobility and fertility.
An interesting question posed by identification of four CatSper genes is how did the CatSper family evolve. Sequence comparison between family members show each CatSper paralogoue to be equally distant from each other, i.e. only around 25% sequence ID. Low sequence identity would argue for an early duplication event or that the CatSper subunits have resulted from convergent evolution of ion channel genes at different chromosomes towards a common function. However, we cannot detect any CatSper like channels in species lower than mouse. This observation would argue for a more recent evolutionary event or rapid evolution. It is notable that sequence identity between repeats within a multi-repeat channel share similar identities to those shared between the CatSper channels ie around 25%. We explored the possibility that a particular CatSper channel would represent one of the four repeating units found in channels such as the L-type calcium channel. However, we cannot form a direct one-to-one relationship between a particular channel repeat and a CatSper unit to support this theory.
CatSper ion-channels present themselves as attractive potential targets for non-hormonal contraceptives. Benoff et al [26] have already illustrated the reversible contraceptive effect of Nifedipine, a widely used calcium-channel blocker in the treatment of high blood pressure and migraine. These effects are mediated via voltage-gated calcium-channels, primarily the L-type voltage-gated channels. The relationship of the CatSper subunits to the voltage-gated calcium channels, their established role in sperm motility and their testis restricted expression pattern, therefore makes them a highly validated target for the identification of novel contraceptives.
Concluding remarks
Based on our identification of two novel CatSper channels and interaction domains we have presented a theoretical model that suggests the CatSper proteins form subunits of a hetero-tetrameric Ca2+ channel in sperm. We also further suggest that experimental determination of this hypothesis and pharmacological studies may lead to the identification of non-hormonal contraceptives.
Author's contributions
AL identified the human and mouse CatSper4 genes and was responsible for the majority of the bioinformatics in the study. VP, LR and LA were responsible for verifying the human CatSper4 transcript and determining the tissue distribution. DM was responsible for identifying human and mouse CatSper3 genes and coordinating the study.
Note added in proof
Human CatSper 3 and CatSper 4 predicted sequences have been submitted to EMBL Nucleotides databases under the accession numbers BN000272 and BN000273 respectively.
Acknowledgments
Acknowledgements
The authors would like to thank John Overington, Richard Fagan, Janet Allen and colleagues at Inpharmatica for helpful comments and discussions.
Contributor Information
Anna Lobley, Email: a.lobley@inpharmatica.co.uk.
Valerie Pierron, Email: v.pierron@inpharmatica.co.uk.
Lindsey Reynolds, Email: l.reynolds@inpharmatica.co.uk.
Liz Allen, Email: l.allen@inpharmatica.co.uk.
David Michalovich, Email: d.michalovich@inpharmatica.co.uk.
References
- Serrano CJ, Trevino CL, Felix R, Darszon A. Voltage-dependent Ca(2+) channel subunit expression and immunolocalization in mouse spermatogenic cells and sperm. FEBS Lett. 1999;462:171–176. doi: 10.1016/S0014-5793(99)01518-5. [DOI] [PubMed] [Google Scholar]
- Westenbroek RE, Babcock DF. Discrete regional distributions suggest diverse functional roles of calcium channel alpha1 subunits in sperm. Dev Biol. 1999;207:457–69. doi: 10.1006/dbio.1998.9172. [DOI] [PubMed] [Google Scholar]
- Wiesner B, Weiner J, Middendorff R, Hagen V, Kaupp UB, Weyand I. Cyclic Nucleotide-gated Channels on the Flagellum Control Ca2+ Entry into Sperm. J Cell Biol. 1998;142:473–484. doi: 10.1083/jcb.142.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel MK, Marrero H, Birnbaumer L, Lemos JR, Florman HM. Trp2 regulates entry of Ca2+ into mouse sperm triggered by egg ZP3. Nat Cell Biol. 2001;3:499–502. doi: 10.1038/35074570. [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, Tilly JL, Clapham DE. A sperm ion channel required for sperm motility and male fertility. Nature. 2001;413:603–9. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quill TA, Ren D, Clapham DE, Garbers DL. A voltage-gated ion channel expressed specifically in spermatozoa. Proc Natl Acad Sci U S A. 2001;98:12527–31. doi: 10.1073/pnas.221454998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers DL. Ion channels. Swimming with sperm. Nature. 2001;413:579. doi: 10.1038/35098164. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Anang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Wise2 :Intelligent algorithms for DNA searches http://www.ebi.ac.uk/Wise2/index.html
- National Centre for Biotechnology Information http://www.ncbi.nlm.nih.gov
- Lobenhofer EK, Bennett L, Cable PL, Li L, Bushel PR, Afshari CA. Regulation of DNA replication fork genes by 17beta-estradiol. Mol Endocrinol. 2002;16:1215–1229. doi: 10.1210/me.16.6.1215. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JALVIEW-A Java Multiple Alignment Editor http://www2.ebi.ac.uk/~michele/jalview/
- Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Adachi J, Aizawa K, Akahira S, Akimura T, Arai A, Aono H, Arakawa T, Bono H, Carninci P, Fukuda S, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Lim S, Jin K, Friedman E. Mirk protein kinase is activated by MKK3 and functions as a transcriptional activator of HNF1alpha. J Biol Chem. 2002;277:25040–25046. doi: 10.1074/jbc.M203257200. [DOI] [PubMed] [Google Scholar]
- ENSEMBL Genome Browser http://www.ensembl.org/
- Stuhmer W, Conti F, Suzuki H, Wang XD, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- Ellinor PT, Yang J, Sather WA, Zhang JF, Tsien RW. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron. 1995;15:1121–32. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, Barnes AA, Emson P, Foord SM, Marshall FH. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:629–630. doi: 10.1038/25248. [DOI] [PubMed] [Google Scholar]
- Kammerer RA, Frank S, Schulthess T, Landwehr R, Lustig A, Engel J. Heterodimerization of a functional GABAB receptor is mediated by parallel coiled-coil alpha-helices. Biochemistry. 1999;38:13263–13269. doi: 10.1021/bi991018t. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–53. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Benoff S. Voltage dependent calcium channels in mammalian spermatozoa. Front Biosci. 1998;3:D1220–40. doi: 10.2741/a358. [DOI] [PubMed] [Google Scholar]