Abstract
The SWI/SNF complex in yeast and Drosophila is thought to facilitate transcriptional activation of specific genes by antagonizing chromatin-mediated transcriptional repression. The mechanism by which it is targeted to specific genes is poorly understood and may involve direct DNA binding and/or interactions with specific or general transcription factors. We have previously purified a mammalian complex by using antibodies against BRG1, a human homologue of SWI2/SNF2. This complex is likely functionally related to the yeast SWI/SNF complex because all five subunits identified so far (referred to as BAFs, for BRG1-associated factors) are homologues of the yeast SWI/SNF subunits. However, we now describe the cloning of the 57-kDa subunit (BAF57), which is present only in higher eukaryotes but not in yeast. BAF57 is shared by all mammalian complexes and contains a high-mobility-group (HMG) domain adjacent to a kinesin-like region. Both recombinant BAF57 and the whole complex bind four-way junction (4WJ) DNA, which is thought to mimic the topology of DNA as it enters or exits the nucleosome. Surprisingly, complexes with mutations in the HMG domain of BAF57 can still bind 4WJ DNA and mediate ATP-dependent nucleosome disruption. Our work describes the first DNA binding subunit for SWI/SNF-like complexes and suggest that the mechanism by which mammalian and Drosophila SWI/SNF-like complexes interact with chromatin may involve recognition of higher-order chromatin structure by two or more DNA binding domains.
Keywords: BRG1-associated factor, chromatin, transcription
The SWI/SNF complex was originally identified in yeast and is thought to facilitate transcriptional activation of several inducible genes by antagonizing chromatin-mediated transcriptional repression (1–5). The complex has been purified (6, 7) and shown to contain an ATP-dependent nucleosome disruption activity. This activity can lead to enhanced binding of transcription factors to nucleosomal templates (7–9). The complex is 2 megadaltons in size (10) and 8 of its 11 subunits have been characterized: SWI1, SWI2/SNF2, SWI3, SNF5, SNF6, SNF11, TFG3, and SWP73 (4, 11–15).
In mammals, several forms of SWI/SNF-like complexes have been partially purified (16) and purified (17) and shown to have similar ATP-dependent nucleosome disruption activities as the yeast SWI/SNF complex (16–19). They seem to be involved in transcriptional activation by the glucocorticoid and retinoid acid receptors (20, 21), in tumor suppression by interacting with retinoblastoma protein (22, 23), and in integration of HIV by interacting with its integrase (24). The mammalian complexes we purified consist of 9–12 subunits depending on the cell line used for purification (17). Five subunits have been cloned: BAF190 (BRG1, hbrm) (20, 21, 25), BAF170 (26), BAF155 (26), BAF60 (26), and BAF47 (17, 24, 27). They are homologues of yeast SWI/SNF subunits, SWI2/SNF2, SWI3, SWI3, SWP73 and SNF5, respectively.
Recently, a complex similar to SWI/SNF has been purified from yeast (28). This complex (called Rsc) is present at much higher level than SWI/SNF. Mutations of several Rsc subunits produce phenotypes that are distinct from SWI/SNF mutations, indicating that its biological functions are also different (28–30). Rsc is required for yeast mitotic growth whereas SWI/SNF is not. The Rsc subunits identified so far (Sth1p, Rsc8, sfh1p, and Rsc6p) are all homologous to the yeast SWI/SNF subunits (SWI2/SNF2, SWI3, SNF5, and SWP73, respectively). The same four Rsc subunits are also homologous to the mammalian BRG1-associated factors (BAFs) (190, 170/155, 47, and 60, respectively). Thus, in terms of the subunit composition, the mammalian complex is as much related Rsc as to SWI/SNF. In terms of abundance, the mammalian complex is more related to Rsc than to SWI/SNF because it is the most abundant form within the cell (17). Furthermore, BRG1 has recently been shown to be required for the viability of F9 murine embryonal carcinoma cells (31). Thus, in terms of function, the mammalian complex may be more similar to Rsc. The name SWI/SNF for the mammalian complex could be misleading, because the mammalian complex could be a functional homologue of the Rsc complex. Therefore, we will refer to the complex as BAF for Brg1 or brm-associated factors.
A fundamental question regarding the mechanism of action of the SWI/SNF complex is how it is targeted to a specific chromosomal locus. At least three models have been proposed from different studies. In one model, the complex is recruited to the promoter through interactions with sequence-specific transcription factors, such as glucocorticoid receptor (32). In the second model, the complex is recruited through stable association with RNA polymerase II holoenzyme (33). A third model was suggested by the observation that the yeast SWI/SNF complex contains intrinsic DNA binding activities similar to high-mobility-group (HMG) proteins (34). All three models may be correct under certain circumstances, and they may not be mutually exclusive. However, which subunit(s) in the complex interacts with sequence-specific transcription factors, associates with pol II holoenzyme or binds DNA is not clear.
We previously demonstrated that a 57-kDa subunit, BAF57, is present in all mammalian BAF complexes that have been purified (17). In contrast, several other subunits are selectively expressed in specific cell types, suggesting that BAF57 is a core component of the mammalian BAF complexes (17, 26). In this study, we report the molecular cloning of the genes encoding BAF57 from human and mouse. We find that the BAF57 sequence contains an HMG domain and a kinesin-like coiled-coil region. The recombinant BAF57 HMG domain as well as the entire mammalian complex can bind DNA with characteristics similar to other HMG proteins. In addition, BAF57 is a mammalian subunit that has no corresponding homologue in the yeast SWI/SNF complex underlining the possibility that the mechanism of the yeast complex may not be strictly applicable to the mammalian complex.
MATERIALS AND METHODS
Purification of Mammalian BAF Complexes and Cloning of BAF57.
The fractionation of nuclear extract and immunopurification of BRG1, hbrm, and BAF60b complexes have been previously described (17, 26). The purified BAF complex from KB cells were separated on 8% SDS gel and transferred to polyvinylidene difluoride (PVDF) membrane. The band corresponding to BAF57 was excised and digested with trypsin. The peptides mixture were fractionated on a reverse-phase HPLC and subsequently sequenced by Edman degradation. The peptide sequences obtained were used to search the NR and dbEST database at the National Center for Biotechnology Information (NCBI) by using the blast program. The sequences were found to match multiple expressed sequence tag (EST) sequences from both human and mouse (access nos. T34541, T36278, D21493). PCR primers were designed based on the EST sequences and were used to amplify the EST DNA by using the cDNA of human Jurkat T cells as the template. The PCR product was then used as a probe to obtain the full-length cDNA as described (26). There is a in-frame stop codon upstream of the predicted initiator metheonine in both human and mouse cDNAs. The sequence alignments were performed by using bestfit, pileup, and pretty of the uwgcg program. Two rabbit polyclonal antibodies were raised against two different regions of BAF57: BAF57H (aa 63–153) and BAF57C (aa 316–411). These regions were fused to maltose-binding protein (New England Biolabs) and the recombinant fusion proteins were expressed and purified according the manufacturer’s protocol and used as immunogen. BAF57 complex was immunopurified with BAF57C antibody by using conditions identical to those for BRG1 complex. The Western blotting conditions are also the same as BRG1 antibodies (17).
Establishment of Cell Lines That Stably Expresses Hemagglutinin (HA)-Tagged BAF57 and Purification of the Complex by Using Antibody Against HA.
The coding region of BAF57 was amplified by PCR to generate appropriate restriction ends and cloned into pBJ5-neo vector that was designed to have a HA epitope at 3′ end of the gene. The plasmid (10 μg) was transfected into Jurkat T cells (1 × 107 cells) by electroporation, and stable transformants were selected with G418 (1 mg/ml). The clones resistant to G418 were screened for expression of HA-tagged BAF57 by Western blot analysis with both 12CA5 mAb and BAF57 antibody. Four independent clones were obtained. Purification of HA-tagged BAF57 complex from three different clones yield identical set of polypeptides as visualized on silver-stained gels. The immunopurification with 12CA5 antibody was similar to that used for purification of HA-tagged TFIID (35). The antibody-protein A beads were prepared as described (17). The crude nuclear extract from the stable cell line (1.3 ml, 7 mg/ml protein concentration) was centrifuged at 100,000 × g to remove cell debris. The supernatant was applied directly to 100 μl of antibody beads and the mixture was rotated at 4°C for 5 h. The beads were then washed four times with 0.5 M buffer D (20 mM Hepes, pH 7.9/0.5 M KCl/0.25 mM EDTA/10% glycerol/1 mM DTT/0.1% Tween-20), once with buffer D lacking KCl, and once with 0.2 M buffer D. The complex was eluted off the beads by incubation at room temperature for 0.5 h with the HA epitope peptide (Anagen) at 1 mg/ml in 0.2 M buffer D.
Expression and Purification of Recombinant BAF57 in Escherichia coli.
BAF57 and its truncated version (containing N-terminal 163 amino acid residues) were expressed with a 6× histidine tag and purified with a Ni2+ column according to the manufacturer’s instructions (Qiagen, Chatsworth, CA). Some contaminating E. coli proteins were removed by heat inactivation at 70°C for 5 min. The recombinant protein was over 95% pure as determined by SDS/PAGE (12%) and used in DNA binding studies. Further purification of the protein was performed by preparative SDS/PAGE. The protein was eluted from the gel and renatured. The renatured protein still retains binding activity to four-way junction (4WJ) DNA. The generation of K112I mutant was done by PCR-mediated mutagenesis. The mutant protein was similarly purified but without the step of preparative SDS/PAGE.
Gel-Shift and Mononucleosome-Disruption Assays.
The 4WJ DNA and its two duplex DNA “arms” were made according to Bianchi (36). The 20 μl reaction mixture for the gel-shift assay contains 0.1 ng p32-labeled probe, 10 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 5% glycerol, 0.1 mM DTT, 0.01% spermidine, 0.1 mg/ml BSA, 300 ng poly(dI-dC), and indicated amount of recombinant protein or complex in the figure legends. The reaction was performed at 4°C for 30 min, and the mixture was analyzed on a 4% native gel (0.5× TBE, the acrylamide to bisacrylamide ratio is 30:1 for recombinant proteins and 80:1 for the BAF57 complex). The electrophoresis was run at 4°C at 10 V/cm. The mononucleosome disruption assay with the 5S DNA as the template was done as described for the yeast complex (8).
RNase Protection Assay for BAF57 mRNA Levels.
The conditions for RNase protection assay is essentially the same as those previously described (37). The BAF57 probe was generated by first cutting pBluscript–mBAF57 cDNA plasmid with BamHI to remove the cDNA from 632 bp to 1,405 bp. The resulting construct was then linearized with BglII and then transcribed with T3 RNA polymerase. The protected fragment should be 220 nucleotides. The actin probe has been described (38).
RESULTS
Purification and Cloning of BAF57.
The BAF57 protein was purified by isolating the BAF complex from human KB cells by using conventional chromatography and immunoaffinity methods (17) (Fig. 1a, lane 2). Five peptide sequences were obtained from microsequencing (see Materials and Methods), which did not match any known genes in the NR databank maintained at NCBI. Based on the peptide sequences, a human cDNA clone was isolated and sequenced. A single ORF was identified containing all five peptide sequences obtained from the pure protein (Fig. 2a).
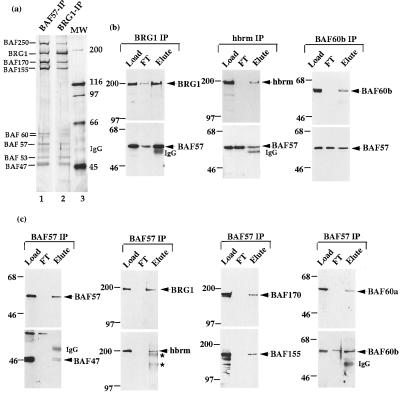
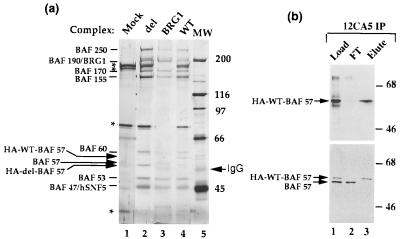
Figure 1.
BAF57 is a shared core subunit of different mammalian BAF complexes. The purified mammalian BAF complex was analyzed by SDS/PAGE (8%) and silver staining (a). The antibodies used for immunopurification are marked on the top of each lane. MW refers to the molecular weight markers. (b and c) The load, flow-through (FT), and eluted fractions from the indicated immunoaffinity columns were analyzed by Western blot analysis by using antibodies indicated at the side of each panel by an arrow. The presence of IgG and crosslinked IgG (∗) were marked.
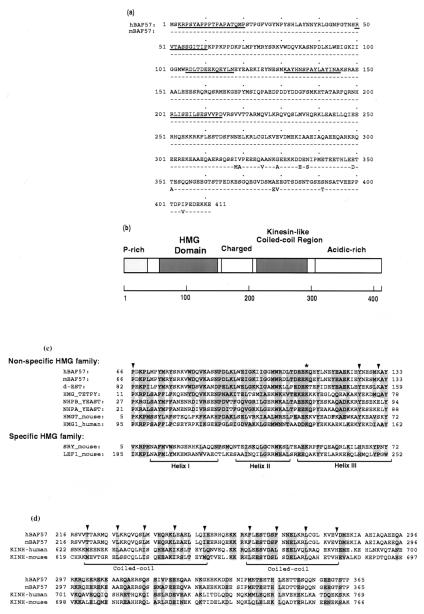
Figure 2.
BAF57 contains a single HMG domain and a coiled-coil region similar to kinesin. (a) The sequence of human and mouse BAF57 proteins are compared, with identical residues represented by dashed lines. The underlined sequences represent peptide sequences obtained from microsequencing tryptic peptides of human BAF57 protein. (b) The domain structures of BAF57 are diagrammed. (c) The HMG domain of BAF57 is aligned with other HMG domains. The arrows indicate amino acid residues that distinguish between specific and nonspecific HMG families. The asterisk indicates the conserved lysine that has been mutated in the experiment described in Fig. 5d. dEST represents an EST sequence (ID AA201846 at dbEST databank maintained at NCBI) that likely encodes the Drosophila homologue of BAF57 (70% identical, 90% similar within the HMG domain). (d) The predicted coiled-coil region of BAF57 is aligned with kinesin. The arrows indicate the hydrophobic residues of the heptad repeats.
BAF57 Sequence Contains an HMG Domain and Kinesin-Like Coiled-Coil Region.
Examination of the predicted sequence of BAF57 reveals that it contains a single HMG domain between amino acids 66 and 133 (Fig. 2 b and c). Previous work has shown that the HMG domain serves as a DNA binding motif (39) for not only the abundant HMG chromosomal proteins, but also specific regulators of transcription and cell differentiation. Current evidence suggest that it plays an “architectural” role during formation of higher-order nucleoprotein complexes (40, 41). The HMG domain-containing proteins have been classified into two families: a sequence-nonspecific family that includes HMG chromosomal proteins, and a sequence-specific family that includes transcription factors LEF1 and SRY (reviewed in ref. 42). The HMG domain of BAF57 belongs to the nonspecific HMG family because amino acid residues corresponding to positions 66, 126, and 131 are all identical to members of this family and are distinct from those of the sequence-specific HMG family. However, proteins of the nonspecific HMG family usually have two or more HMG domains, whereas those of the specific family usually have a single HMG domain. In this later regard, BAF57 is more similar to the specific family. The BAF57 sequence also contains a region with two heptad repeats similar to the coiled-coil region of kinesin (Fig. 2d). The sequence similarity to kinesin extends beyond those residues necessary for the leucine zipper structure, suggesting a similar function. A region rich in acidic residues (net negative charge −28) was found near its C terminus (Fig. 2b), which is similar to the HMG1 protein that also contains a highly negatively charged C terminus.
BAF57 Homologues Are Present in Higher Eukaryotes but Not Yeast.
The mouse homologue of BAF57 was isolated by using the human cDNA as the probe. Comparison of the two sequences revealed that they are highly similar to each other (97% identical, 99% similar at amino acid level) and their HMG domains are identical. A partial Drosophila cDNA sequence has been found in dbEST databank which shows significant homology to human BAF57 (70% identity and 90% similarity within the HMG domain) (Fig. 2c). Preliminary data suggest that this cDNA encodes the homologous subunit of BAF57 within the Drosophila SWI/SNF-like complex (G. Daubresse, W.W., and M. Scott, unpublished data). We could not find any ORFs that have significant homology to BAF57 outside the HMG domain from the completed genome database for Sacchromyces cerevisiae. None of the yeast SWI/SNF subunits identified so far contains a discernible HMG domain. The remaining three uncharacterized subunits of the yeast SWI/SNF complex do not contain any HMG domains (B. Cairns, personal communication). Thus, BAF57 is present only in SWI/SNF-like complexes from higher eukaryotes but not yeast, underscoring the differences between these complexes.
BAF57 Is a Core Component Shared by Different Mammalian SWI/SNF-Like Complexes.
Two rabbit antibodies were made against BAF57 by using two different regions of the predicted ORF as immunogens (Materials and Methods). Both antibodies recognize the 57-kDa subunit in the BRG1 complex by Western blot analysis (Fig. 1b Left; data not shown), indicating that the isolated cDNA does encode BAF57. The antibodies also recognize the 57-kDa component of complexes purified with antibodies against hbrm (17) and BAF60b (26) (Fig. 1b Middle and Right; data not shown). Western blot analysis the polypeptides purified with the BAF57 antibody column revealed the presence of every known mammalian BAF subunit (Fig. 1b; data not shown). The pattern of these BAF57-associated polypeptides on a silver-stained SDS gel is remarkably similar to those associated with the BRG1 complex (Fig. 1a, compare lanes 1 and 2), indicating that BAF57 is a core subunit shared among different mammalian BAF complexes.
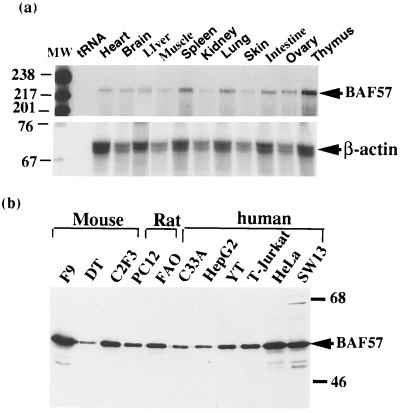
BAF57 mRNA was detected in all tissue examined at about equal levels (Fig. 3a). The BAF57 protein was also found in cell lines derived from several different tissues by Western blot analysis (Fig. 3b). Thus, BAF57 appears to be ubiquitously expressed, similar to other core subunits of the complex [BAF170, BAF155, and BAF47/hSNF5 (26)].
Figure 3.
BAF57 is widely expressed in different tissues and cell lines. (a) RNA from different mouse tissues were examined by quantitative RNase protection analysis (35). The tissue source for the RNA and the tRNA control are indicated on the top of each lane. The probe for actin mRNA is used to normalize the RNA loaded for each lane (Lower). The RNA products were separated on a 6% polyacrylamide-urea gel and autoradiographed by using x-ray films. (b) Nuclear extracts from several cell lines derived from different tissue sources were analyzed by Western blot analysis by using anti-BAF57 antibodies. The name of each cell line and its species is indicated on the top of each lane and their description can be found in ref. 17.
The Recombinant BAF57 HMG Domain and the Mammalian BAF Complex Bind 4WJ DNA.
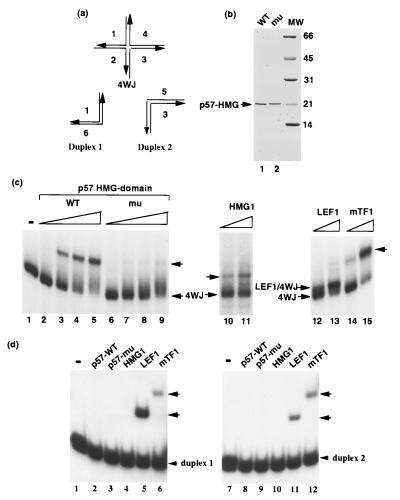
A unique feature of the HMG domains is their high affinity for DNA with sharp bends such as 4WJ DNA and low affinity for duplex DNA (43) (Fig. 4a). We investigated the DNA binding properties of BAF57 by expressing it as a recombinant protein in E. coli. The full-length protein was expressed at very low levels and has many degradation and/or premature termination products. A truncated protein containing the entire HMG domain was found stably expressed at high levels and was purified to homogeneity (Fig. 4b). This protein shows strong binding activity to 4WJ DNA in gel-shift assays (Fig. 4c, and data not shown), which is believed to mimic the entry and exit points from the nucleosome (44). As a control, a point mutant was created by changing a conserved lysine residue within the HMG domain to isoleucine (K112I, Fig. 2c). The mutant protein was purified (Fig. 4b, lane 2), and showed at least 10-fold lower DNA binding activity compared with the wild type (Fig. 4c, lanes 6–9). An identical mutation has previously been described in the HMG domain of SRY that similarly decreased the DNA binding activity (45). When the duplex “arms” of the 4WJ DNA (depicted in Fig. 4a) were used as probes, the recombinant BAF57 proteins showed no detectable binding activity even at a concentration 10-fold higher than those necessary to bind 4WJ DNA (Fig. 4c, lanes 6–9). These DNA binding properties of BAF57 are similar to the HMG1 protein and the yeast SWI/SNF complex (34), but different from two other HMG domain-containing proteins, LEF1 and mTF1 (Fig. 4 c and d). In addition, BAF57 showed no detectable binding to 12 other duplex probes of different sequences and lengths (data not shown). Our attempts to define a consensus binding site for BAF57 by using several binding site selection methods were unsuccessful. Thus, BAF57 appears to be highly selective for 4WJ DNA as is the yeast SWI/SNF complex (34).
Figure 4.
The BAF57 HMG domain binds 4WJ DNA similar to the HMG1 protein. The probes illustrated in a are used to examine the binding properties of the mammalian SWI/SNF complexes and recombinant BAF57. The diagrams illustrate the structures of the 4WJ DNA and the two regular duplex DNA that made up its “arms.” (b) The truncated recombinant BAF57 proteins purified from E. coli were analyzed on a Coomassie blue-stained SDS gel (12%). (c) The DNA binding activities of wild-type and K112I mutant BAF57 proteins were analyzed by gel-shift assays by using 4WJ DNA as the probe. The concentrations of proteins are 30, 100, 300, and 1,000 nM, respectively. Also shown in the figure are binding of three other HMG box-containing proteins: HMG1, LEF1, and mTF1. Their concentrations are 300 and 1000 nM, respectively. (d) The DNA binding activities of recombinant BAF57 proteins were analyzed by gel-shift assays by using two duplex “arms” of 4WJ DNA as probes. The protein concentration is 1,000 nM for all proteins. Note that the recombinant proteins may not be 100% active. The arrows indicate the DNA–protein complex formed.
To develop a way of studying the DNA binding characteristics of complexes having mutant BAF57 proteins, we established a stable cell line expressing BAF57 tagged with the HA epitope and purified the BAF complex with a monoclonal antibody against HA (Fig. 5 a, lane 4, and b). The use of the mAb (12CA5) column allows the elution of the complex under near physiological salt conditions by using the HA peptide. The strategy has been used previously to purify the holo-TFIID complex (35). The HA-tagged exogenous BAF57 has a slower mobility on SDS gel compared with the endogenous BAF57, caused by the molecular weight increase by addition of the HA tag (Fig. 5b; also compare lanes 3 and 4 of Fig. 5a). This mobility difference provides a convenient way to distinguish the exogenous and endogenous proteins. The BAF complex purified with the anti-HA antibody contained only the HA-tagged BAF57, but not the endogenous BAF57. Because the level of the tagged protein in the cell is lower than that of the endogenous protein, the result suggests that there is only one BAF57 per complex (otherwise, the endogenous BAF57 will also be present in the complex purified with anti-HA antibody). This result contrasts to results with the SWI3-like subunits, BAF170 and BAF155, which are present in two copies per complex (26).
Figure 5.
Purification of BAF complex containing exogenously introduced BAF57 tagged with the HA epitope. (a) The subunit composition of the HA-tagged BAF57 complexes purified by 12CA5 mAb is shown on a silver-stained SDS gel (8%). The complex containing the wild-type (WT) or the deletion mutant (del) are marked on the top of each lane. Mock refers to the control experiment by using the parental cell line without the exogenous BAF57 gene as the source for 12CA5 immunopurification. BRG1 represents the complex purified with BRG1 antibody. Several contaminating proteins were marked by ∗. The arrows indicate the HA-tagged wild-type BAF57 (WT), the deletion mutant whose entire HMG domain was removed (del), and the endogenous BAF57. They showed the different mobilities on the SDS gel caused by the addition of the HA tag. (b) The load, flow-through (FT), and elute fractions of 12CA5 immunoaffinity column for purification of the complex containing the HA-tagged wild-type BAF57 were analyzed by Western blotting. The arrows indicate the presence of immunoreactivity to 12CA5 antibody (Upper) or BAF57 antibody (Lower).
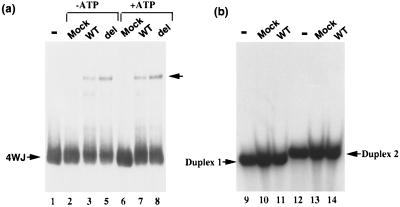
The BAF complex purified from the cells stably expressing the HA-tagged BAF57 was tested for its DNA binding properties by using the gel-shift assays and probes described above. This complex shows clear binding activity to 4WJ DNA (Fig. 6a), but no detectable activity to regular duplex DNA (Fig. 6b). The presence or absence of ATP in the reaction does not change the binding (Fig. 6a). Thus the mammalian BAF complex has similar DNA binding characteristics as the BAF57 HMG domain and the yeast SWI/SNF complex.
Figure 6.
The mammalian BAF complex binds 4WJ DNA, a property similar to the yeast complex and HMG proteins. The DNA binding activity of HA-tagged BAF57 complex is analyzed by gel-shift assays by using either 4WJ DNA (a) or its two duplex DNA “arms” (b), as indicated by the arrows. The concentration of the complex is 5 nM. The complex containing either the HA-tagged wild-type BAF57 (WT) or the HMG deletion mutant (del) are indicated on the top of each lane. Mock refers to the negative control as described in the legend to Fig. 5. The presence or absence of 1 mM ATP is also indicated.
The BAF57 HMG Domain Is Not Essential for the DNA Binding Activity or the Nucleosome Disruption Activity of the BAF Complex.
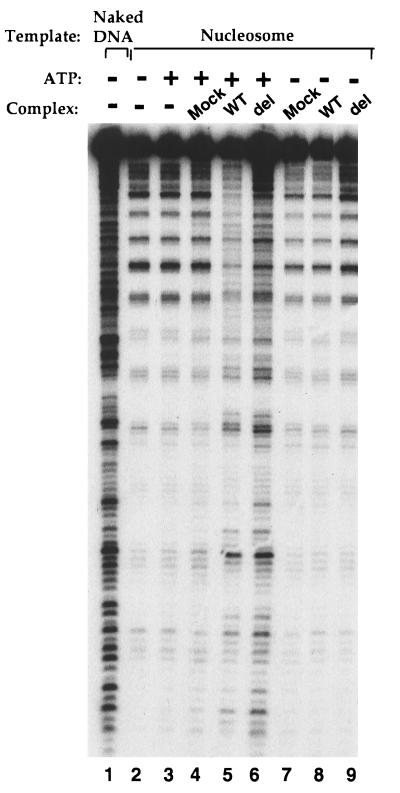
We investigated the importance of the HMG domain in the context of the whole complex by using the above in vivo assembly method. We constructed two additional stable cell lines expressing HA-tagged BAF57 mutants: the same K112I mutation within the HMG domain that has been shown to decrease the binding affinity to 4WJ DNA by recombinant BAF57 by at least 10-fold (Fig. 4c), and a deletion mutant that removes the entire HMG domain (aa 62–131). Both mutant complexes were purified with antibody against HA and found to contain all other subunits (Fig. 5a, compare lanes 2 and 4; data not shown for the point mutant), indicating that the HMG domain is not involved in assembly of the complex (the N-terminal proline-rich region is important for assembly of the complex; unpublished data). Surprisingly, both mutant complexes have 4WJ DNA binding activity and ATP-dependent nucleosome disruption activity indistinguishable from the wild-type complex (Figs. 6a and Fig. 7; data not shown for the point mutant), suggesting the HMG domain is not essential or is redundant for these in vitro activities of the mammalian BAF complex.
Figure 7.
The HMG domain of BAF57 is not essential for the in vitro nucleosome-disruption activity of the complex. The BAF complexes containing either the wild-type (WT) or the deletion mutant (del) of BAF57, as indicated on the top of the figure, were analyzed for the ATP-dependent nucleosome disruption activities. The naked DNA template and the nucleosomal template are shown on the top of the figure. The presence or absence of 1 mM ATP is also indicated.
DISCUSSION
We report the cloning of BAF57, a subunit of mammalian SWI/SNF-like complexes (referred to as BAF). At least three lines of evidence indicate that BAF57 is a component of all mammalian BAF complexes. First, BAF57 stably associates with all other BAF subunits even under highly stringent conditions, such as 1 M KCl or 0.2 M guanidine hydrochloride. Second, immunopurification with any existing BAF antibody copurifies BAF57. Third, complexes purified with the BAF57 antibody have similar subunit compositions as those purified by anti-BRG1, -hbrm, or -BAF60b antibodies (Fig. 1a) (17, 26). Because yeast do not have a homologue of BAF57, our data provide evidence of the structural differences between the mammalian BAF and yeast SWI/SNF-like complexes. Our preliminary data suggest that at least two other BAF subunits do not have yeast homologues and that several yeast SWI/SNF subunits do not have homologues in the BAF complex (unpublished data). Thus, although the BAF complex is similar to the yeast SWI/SNF and Rsc in several regards, it has significant differences in overall structure and possibly biological functions.
BAF57 Contains an HMG-Like DNA Binding Domain that Belongs to the Nonspecific HMG Family of Proteins.
HMG domains have been found in the abundant chromosomal proteins HMG1/2 as well as specific transcription factors such as UBF, LEF1, and SRY. They interact with minor grooves of DNA and can induce strong bending on association with their templates (40, 41, 46–48). Recent evidence suggests that they may not have biological activities on their own, but rather function as architectural factors during assembly of higher-order nucleoprotein complexes with other proteins (reviewed in ref. 42). The fact that the BAF complex contains an HMG domain-containing subunit implies that the complex may play an architectural role like other HMG domains. We demonstrated that the recombinant BAF57 HMG domain binds DNA similar to HMG proteins, with high affinity to 4WJ DNA and little affinity to linear DNA. To our knowledge, this is the first DNA binding subunit identified for the SWI/SNF-like complexes from any species. The yeast SWI/SNF complex has been shown to bind 4WJ DNA like HMG proteins (34), yet none of the subunits from the yeast complex has been shown to have DNA binding characteristics of HMG proteins.
The Mammalian BAF Complex Contains Additional DNA Binding Domains Other than BAF57.
We showed that the BAF complex has similar DNA binding characteristics as the recombinant HMG domain of BAF57 and the yeast SWI/SNF complex. They all have high affinity to 4WJ DNA and low affinity to linear DNA, indicating that the DNA binding properties of SWI/SNF-like complexes are conserved through evolution. However, the domain through which the BAF complex interacts with DNA or nucleosomes may not necessarily be BAF57, because the BAF complex containing HMG domain-deleted BAF57 still has intact DNA binding activity (Fig. 6) and the ATP-dependent nucleosome disruption activity (Fig. 7). Thus, the BAF complex must contain other DNA binding subunit(s). Without the HMG domain of BAF57, the complex is able to bind 4WJ DNA and disrupt nucleosomes through these other subunit(s).
What, then, is the role for the BAF57 HMG domain within the mammalian BAF complex? Because the yeast SWI/SNF complex has similar DNA binding properties as the BAF complex and it does not have a BAF57 homologue, the primary interactions of both complexes to DNA should be through subunit(s) other than BAF57. The interaction provided by BAF57 HMG domain to its target could be secondary or redundant for the mammalian complex. As a result, the mammalian complex may be able to interact with multiple nucleosomes and structured DNA through two or more DNA binding domains. The interaction through BAF57 may provide additional specificity during target recognition by the BAF complex. Because the intracellular concentrations of BAF in mammals or SWI/SNF in yeast relative to their DNA targets is not known, the in vitro binding studies reported here and for yeast (34) must be viewed with some skepticism.
Alternatively, BAF57 may not be involved in targeting at all but is used at some stage in the disassembly of chromatin from higher-order structure to a less compact structure. Because the DNA binding characteristics of the yeast and the mammalian SWI/SNF-like complexes have been studied on bare DNA, the role for the HMG domain of BAF57 may not be discernible with the available assays. Identification of the other DNA binding subunit(s) will help to elucidate the importance of BAF57 during DNA binding. Studies of the complex bearing mutant BAF57 proteins during development may be necessary to discern its biological functions.
Acknowledgments
We thank Dr. R. Tjian for his support and advice throughout this project. We also thank Drs. Hui Ge, R. Johnson, and S. Lippard for providing purified recombinant HMG1 protein; Dr. C. Pabo for LEF1; and Dr. D. Clayton for mTF1. We also want to thank Drs. Jacques Cote and Tom Hughes for advice and materials for nucleosome disruption assays. W.W. has been previously supported by fellowships from Damon Runyon–Walter Winchell Cancer Foundation (DRG-1182) and the Howard Hughes Medical Institute.
ABBREVIATIONS
- BAF
BRG1-associated factor
- HMG
high mobility group
- 4WJ
four-way junction
- EST
expressed sequence tag
- HA
hemagglutinin
- NCBI
National Center for Biotechnology Information
Footnotes
References
- 1.Stern M, Jensen R, Herskowitz I. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 2.Neigeborn L, Carlson M. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasmyth K, Stillman D, Kipling D. Cell. 1987;48:579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 4.Peterson C L, Herskowitz I. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 5.Hirschhorn J N, Brown S A, Clark C D, Winston F. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 8.Owen-Hughes T, Utley R T, Cote J, Peterson C L, Workman J L. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 9.Utley R T, Cote J, Owen-Hughes T, Workman J L. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- 10.Peterson C L, Dingwall A, Scott M P. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurent B C, Treitel M A, Carlson M. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estruch F, Carlson M. Mol Cell Biol. 1990;10:2544–2553. doi: 10.1128/mcb.10.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treich I, Cairns B R, de los Santos T, Brewster E, Carlson M. Mol Cell Biol. 1995;15:4240–4248. doi: 10.1128/mcb.15.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cairns B R, Henry N L, Kornberg R D. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 16.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 18.Imbalzano A N, Kwon H, Green M R, Kingston R E. Nature (London) 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 19.Imbalzano A N, Schnitzler G R, Kingston R E. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 20.Muchardt C, Yaniv M. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiba H, Muramatsu M, Nomoto A, Kato H. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 23.Singh P, Coe J, Hong W. Nature (London) 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 24.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 25.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. Nature (London) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Xue Y, Zhou S, Kuo A, Cairns B, Crabtree G R. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 27.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 29.Treich I, Carlson M. Mol Cell Biol. 1997;17:1768–1775. doi: 10.1128/mcb.17.4.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 33.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 34.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. Nature (London) 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 36.Bianchi M E. EMBO J. 1988;7:843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller C L, Feldhaus A L, Rooney J W, Rhodes L D, Sibley C H, Singh H. Mol Cell Biol. 1991;11:4885–4894. doi: 10.1128/mcb.11.10.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jantzen H M, Admon A, Bell S P, Tjian R. Nature (London) 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 40.Giese K, Cox J, Grosschedl R. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 41.Paull T T, Haykinson M J, Johnson R C. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 42.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 43.Bianchi M E, Beltrame M, Paonessa G. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 44.Lilley D M. Nature (London) 1992;357:282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- 45.Pontiggia A, Rimini R, Harley V R, Goodfellow P N, Lovell-Badge R, Bianchi M E. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van de Wetering M, Clevers H. EMBO J. 1997;11:3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrari S, Harley V R, Pontiggia A, Goodfellow P N, Lovell-Badge R, Bianchi M E. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pil P M, Chow C S, Lippard S J. Proc Natl Acad Sci USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]