Abstract
Polymorphisms in the LRP5 gene have been associated with bone mineral density (BMD) in men and/or women. However, the functional basis for this association remains obscure. We hypothesized that LRP5 alleles could modulate Wnt signaling and the relationship between physical activity and BMD.
This genetic association study was performed in the population-based Framingham Study Offspring Cohort, and included a subset of 1797 unrelated individuals who provided blood samples for DNA and who had BMD measurements of the hip and spine. Ten single-nucleotide polymorphisms (SNPs) spanning the LRP5 gene were genotyped and used for association and interaction analyses with BMD by regression methods. LRP5 haplotypes were transiently co-expressed with Wnt3a, MesD and Dkk1 in HEK293 cells and their activity evaluated by the TCF-Lef reporter assay.
Six out of ten SNPs in LRP5 were associated with one or more of the femur or spine BMDs in men or women after adjustment for covariates, and these associations differed between genders. In men ≤age 60 years, 3 SNPs were significantly associated with BMD: rs2306862 on Exon 10 with femoral neck BMD (p=0.01) and Ward’s BMD (p=0.01); rs4988321/p. V667M with Ward’s BMD (p=0.02); and intronic rs901825 with trochanter BMD (p=0.03). In women, 3 SNPs in intron 2 were significantly associated with BMD: rs4988330 for trochanter (p=0.01) and spine BMD (p=0.003); rs312778 with femoral neck BMD (p=0.05); and rs4988331 with spine BMD (p=0.04). For each additional rare allele, BMD changed by 3–5% in males and 2–4% in females. Moreover, there was a significant interaction between physical activity and rs2306862 in exon 10 (p for interaction=0.02) and rs3736228/p. A1330V in exon 18 (p for interaction=0.05) on spine BMD in men. In both cases, the TT genotype was associated with lower BMD in men with higher physical activity scores, conversely with higher BMD in men with lower physical activity scores. In vitro, TCF-Lef activity in presence of Wnt3a was significantly reduced in cells expressing LRP5 haplotypes carrying the T allele of exon 10 and 18 compared to the wild-type allele, whereas co-expression of Dkk1 completely inhibited Wnt3a response through all LRP5 haplotypes.
In summary, genetic variation in exons 10 and 18 of the LRP5 gene modulates Wnt signaling and the relationship between physical activity and BMD in men. These observations suggest that Wnt-LRP5 may play a role in the adaptation of bone to mechanical load in humans, and may explain some gender-related differences in bone mass.
INTRODUCTION
Bone mineral density (BMD), a major risk factor for osteoporotic fractures, has high heritability.1, 2 Despite ample suggestion from humans and mice that osteoporosis genes may partly differ between males and females,3–5 most association studies with bone density, turnover and/or fractures have focused on gene polymorphisms in women.6–10 The few studies that have examined osteoporosis genes in men, such as the aromatase gene (CYP19)11 and the vitamin D receptor gene (VDR),12 have rarely studied women from the same population. Not only genes, but also environment/life style factors and their interaction with genetic variation play a prominent role in osteoporosis risk;13,14 yet the contribution of interactions between non-genetic and genetic factors remains poorly understood. Among these environmental factors, physical exercise is an important determinant of peak bone mass acquisition and maintenance, which effects may also differ between men and women.15 The mechanisms for gender-related differences in the skeletal response to physical activity are currently unknown. Nevertheless, there is some evidence that genetic variation may influence the response of muscles and the skeleton to intense physical training.16, 17
Recently, mutations in the lipoprotein receptor-related protein 5 (LRP5) gene have been demonstrated to result in different single gene disorders with either a high or low BMD phenotype18. Thus in the autosomal recessive disorder, osteoporosis-pseudoglioma syndrome (OPPG), very low bone mass and blindness have been linked to loss-of-function mutations in the LRP5 gene.19 Concurrently a high bone mass phenotype was described in several extended pedigrees as being linked to gain-of-function mutations in the LRP5 gene.20–22 The high bone mass phenotype is characterized by greater amounts of both cortical and trabecular bone, and a skeleton of greater strength resulting in a complete absence of fractures in one of the pedigrees described.23
LRP5 mutations affect bone formation by osteoblasts, principally by altering Wnt signaling through the canonical beta-catenin pathway.18 In turn, ablation of the LRP5 gene in mice mimics the skeletal phenotypes reported in humans with OPPG.24 Conversely, transgenic mice containing the high bone mass mutation have greater bone mass, density, and strength as compared to non-transgenic littermates.25 Some have suggested that the superior resistance to fracture observed in the transgenic animals may indicate a role for LRP5 in bone adaptation to mechanical load.25, 26
Based on these provocative findings in several human kindreds and in transgenic mice, the contribution of the LRP5 gene to BMD in the general population has recently been investigated.27, 28 In one recent cross-sectional study of 889 healthy Caucasian male and female, significant associations were found for a missense substitution in exon 9 (p.V667M/rs4988321) and its haplotypes with a missense SNP in exon 18 (p.A1330V/rs3736228) with lumbar spine bone mineral content (BMC) and projected bone area.28 The associations with peak bone mass were observed mainly in men, in whom LRP5 polymorphisms accounted for up to 15% of the traits’ variances, and with BMC gain in prepubertal male children. Others have observed a stronger association and linkage between spine BMD, projected bone area and/or fragility fractures and certain alleles of the LRP5 gene in males than in females,29, 30 as well as an association between LRP5 alleles and idiopathic male osteoporosis.31 It is possible therefore that the LRP5 gene could display gender-specific effects on osteoporosis risk. Previous studies, however, did not explore the possibility of interaction between variants in this gene, physical activity, and BMD, nor did they assess the functional significance of LRP5 genetic variation on Wnt signaling.
The Framingham Osteoporosis Study offers a unique opportunity to examine multiple polymorphisms in osteoporosis candidate genes as they relate to BMD at both the spine and hip in a large number of men and women sharing a similar genetic and environmental background32. These observations prompted us to hypothesize that an interaction of physical activity with LRP5 alleles might explain some gender-related differences in bone mass.18
METHODS
Study Sample
In 1971, the Framingham Offspring Study was initiated with the intent to evaluate, among other goals, the role of genetic factors in the etiology of coronary artery disease, and included a total sample size of 5124 subjects.33 The Offspring Cohort is comprised of adult offspring (and their spouses) of couples from the Original Framingham Cohort and 96.4% of the Offspring Cohort are Caucasians, with origin in Eastern and Western Europe. After their initial evaluation, these individuals have undergone repeat examinations approximately every four years, and participated in the Framingham Osteoporosis Study at either their examination cycle 6 or 7, as described in details elsewhere.32, 34 Informed consent was obtained from participants before entry into the study, which was approved by the Boston University Institutional Review Board for Human Subjects Research, and the Hebrew Rehabilitation Center for Aged Institutional Review Board.
Participants in the current study are a subset of unrelated individuals from the Framingham Offspring cohort who provided blood samples for DNA and who had BMD measurements of the hip and spine; 1797 of these (868 men and 929 women) also had genotyping done on most of the ten LRP5 SNPs that were used for the current study. These subjects were not selected on the basis of any trait, but only on the basis that they were biologically unrelated and had DNA.
Bone Mineral Density
The participants underwent bone densitometry of the spine, femoral neck, trochanter, and Ward’s area by dual x-ray absorptiometry using a Lunar DPX-L device to measure BMD (g/cm2) in 1996 – 2001. The coefficients of variation in normal subjects for the DPX-L were 0.9% (spine), 1.7% (femoral neck), 2.5% (trochanter), and 4.1% (Ward’s area).35
Physical Activity
Physical activity was examined using the Physical Activity Scale for Elderly (PASE) questionnaire,36 which has been validated against activity diaries,37 activity monitors,38 and doubly labelled water.39
Other variables
Variables influencing BMD were obtained at the time of BMD measurement along with overall medical history. Details of these measurements have been published previously 35, 40. These variables included age, sex, weight, and for women, estrogen use and menopausal status. Age squared was considered in the models to account for potential non-linear age effects41. Weight was measured using a standardized balance beam scale.35, 40 Analyses were conducted separately by sex and then women were further stratified by menopausal status, where menopause was defined as having no menstrual period for at least one year or no menstrual history for less than a year but taking postmenopausal estrogen. For postmenopausal women, oral estrogen use was evaluated as current use (yes/no). Past users of estrogen replacement therapy (ERT) represented only 0.6% of all women, and were included among never users in all analyses.
DNA Extraction and Genotyping
Genomic DNA was extracted from peripheral blood leukocytes, from Offspring Cohort participants attending their sixth examination cycle (1993–1998), using standard methods. We selected tag SNPs to account for at least 80% of the common haplotypes42 for each of three linkage disequilibrium (LD) blocks in LRP5 previously identified by Twells, et. al.43 In addition, exonic SNPs were selected in the genomic region separating the second and third LD block. From the 13 SNPs initially selected, 3 failed to be genotyped for technical reasons, mostly pertaining to assay design issues. Genotyping was performed using the Applied Biosystems TaqMan SNP genotyping assays, available either on-demand or custom designed. The thermal cycler conditions used for PCR were: denaturation at 95 C for 10 min, 48 cycles of denaturation at 95 C for 15 sec and annealing/extension at 60 C for 1 min. ABI PRISM 7900HT Sequence Detection System was used for allelic discrimination.
In vitro activity of LRP5 haplotypes
We used the Lef-TCF sensitive transcription assay to evaluate canonical Wnt3a signaling mediated by common LRP5 haplotypes. HEK293T were seeded at a density of 8. 104 cells/well in 24-well tissue culture plates in 0.5 ml of complete DMEM and transfected after 24 hours using Fugene® reagent (Roche, Basel, Switzerland) with 100 ng DNA of TOPFLASH luciferase reporter (Upstate, Lucerna, Switzerland) and LRP5 constructs carrying SNPs in exon 9, 10 15 and 18 (haplotype 0, wild-type, G-C-A-C; haplotype 1, G-T-G-T; haplotype 2, A-T-G-T). Cells were co-transfected with expression vectors encoding Wnt3a, MesD-C2 and Dkk1 (generous gifts from Dr. G. Rawadi, Prostrakan, Paris, FR) or empty pcDNA3.1 vector as needed to make the total transfected DNA amount equal in all wells (equivalent to 100ng cDNA for LRP5, Wnt3a and MesD, and 5ng for Dkk1). As an appropriate control for transfection efficiency, HEK cells were co-transfected with plasmid pRLTK, encoding for Renilla luciferase (5 ng DNA per ). Eight hours after transfection, the medium was replaced by fresh medium containing 1% FCS. 24h after transfection, cells were lysed in 300l passive lysis buffer (Promega, Madison, WI). The firefly luciferase activity (Topflash) and the Renilla luciferase activity were quantitated on a TD-20/20 luminometer (Turner BioSystems, Sunnyvale, CA) using the dual Luciferase Assay kit (Promega, Madison, WI) and the specific TCF-Lef reporter activity expressed as relative luciferase units.
Statistical Analysis
The observed SNP genotype frequencies were analyzed, using a 1-degree of freedom χ2 test, to determine if they were in Hardy-Weinberg equilibrium. For each pair of markers linkage disequilibrium was evaluated using D′ and r2 calculated from estimated haplotype frequencies.44 Genotype-phenotype association analyses were conducted separately in men and women to examine the main effects of genotype. Initially genotype groups were compared with regard to covariates using analysis of variance for continuous variables and χ2 statistics for categorical variables.
We assessed the association between polymorphisms in the LRP5 gene and BMD at all bone sites using multiple linear regression, with simultaneous adjustment for covariates. For SNPs with minor allele frequencies of 20% or greater, the effect of the genotypes on BMD was assessed coding each genotype as a factor with three groups (2 degrees of freedom test). For SNPs with minor allele frequency less than 20%, the number of homozygotes for the minor alleles was small, and we coded the genotype as the number of minor alleles (0, 1, or 2) and used additive models for the genotype effect (1 degree of freedom test). Previous work has suggested that 1) LRP5 genetic variation influences peak bone mass acquisition 28 and 2) the strength of the association for some LRP5 SNPs could differ in males and females 28, 29 Since our sample included both genders and was middle-aged, we hypothesized a priori that our findings might be more significant in men and for younger aged members of our sample. Accordingly, analyses were performed separately in men and women, in pre- and postmenopausal women, and in younger (≤ 60 years of age) and older (>60 years of age) men. Age 60 years old was chosen to create a group of men who were young enough not to have manifest the more significant bone loss that comes with advanced age, while providing sufficient sample size for analyses. We generated plots of the −log10 of the p-values for the analyses for each SNP at the various skeletal sites for each of the individual samples such as women and men, young men, and premenopausal women. For SNPs found to be significantly associated with BMD, the proportion of variance in BMD explained by the individual SNP was determined. To estimate the magnitude of effect of SNPs that were significantly associated with BMD, we also compared differences in BMD according to the number of rare alleles. All 10 SNPs were entered into a single model adjusting for all covariates only to estimate the proportion of variance in BMD explained by the set of ten SNPs.
We also performed haplotype analyses using the software haplo.score.45 The haplo.score function estimates haplotype frequencies via the expectation-maximization algorithm and performs statistical tests of association using score statistics for quantitative traits adjusting for the same covariates as included in the single SNP analyses. We performed haplotype analysis for each previously described block separately, and for the exonic SNPs between blocks 2 and 3.43 Haplotype analyses were performed for males and females separately.
Since our main goal was to test for an interaction between physical activity and polymorphisms in the LRP5 gene, we added interaction terms (PASE score*genotype) to the regression models. We also examined the adjusted BMDs according to percentiles of PASE scores across genotype groups for several SNPs for which significant interaction terms were observed (p<0.05). Analyses were performed using R – Language and environment for statistical computing and graphics. (www.r-project.org).
RESULTS
The 10 LRP5 SNPs used in this study are shown in Table 1 with detailed information about each polymorphism, and Figure 1 shows them in the context of the gene structure, as well as linkage disequilibrium (LD) for each pair of markers using D′ and r2 calculated from estimated haplotype frequencies.44 As indicated, SNPs were chosen to span the entire gene locus. The characteristics of the sample of men and women who had both BMD measures and genotyping data are presented in Table 2. The mean ages were in the early sixties, 86% of the women were menopausal and 34% used estrogen (including 0.6% of past users). Smoking was uncommon in both men and women but men tended to be more physically active as measured using the Physical Activity Scale for the Elderly (PASE). In the regression models with all covariates, the proportion of variance (R2) in BMD explained by the covariates varied between 8.5% (spine) and 19.9% (femoral neck) in men. Moreover, in men 60 years of age and younger (n=386), we found a consistent pattern of statistically significant association between physical activity and BMD at almost every skeletal site (increase to R2 by adding PASE to the models ranged from 1.5% to 1.8% with p-values between 0.005 and 0.009). In women, the covariates explained between 24.1% (spine) and 39.5% (trochanter) of the variability in BMD, although physical activity did not contribute significantly to the overall R2.
Table 1.
LRP5 single nucleotide polymorphisms genotyped in this study
| SNP | Location | LD block* | Position** | Alleles | Minor Allele Frequency | |
|---|---|---|---|---|---|---|
| 1 | rs4988330 | Intron 2 | Block 1 | 13386794 | C>T | 7.6% |
| 2 | rs312016 | Intron 2 | Block 1 | 13388198 | A>G | 29.8% |
| 3 | rs4988331 | Intron 2 | Block 1 | 13391128 | T>C | 9.5% |
| 4 | rs312778 | Intron 2 | Block 2 | 13414127 | C>T | 42.4% |
| 5 | rs4988321 | Exon 9 | Inter-block 2 & 3 | 13479984 | A>G † | 5.5% |
| 6 | rs2306862 | Exon 10 | Inter-block 2 & 3 | 13483305 | C>T | 15.4% |
| 7 | rs556442 | Exon 15 | Inter-block 2 & 3 | 13498485 | A>G | 34.5% |
| 8 | rs607887 | Intron 16 | Block 3 | 13502756 | C>T | 34.0% |
| 9 | rs3736228 | Exon 18 | Block 3 | 13507090 | C>T † | 13.1% |
| 10 | rs901825 | Intron 21 | Block 3 | 13511992 | C>G | 10.4% |
LD blocks according to Twells41
position relative to contig number NT_033903.7
Missense substitutions in exon 9 (p. V667M) and 18 (p.A1330V)
Figure 1.
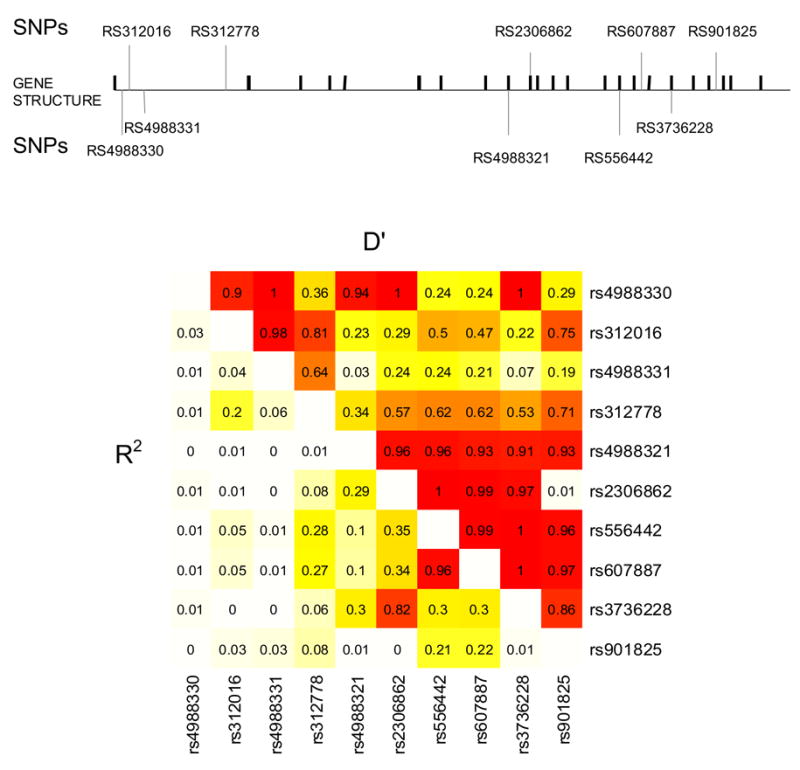
Diagram of the physical map of the LRP5 gene with exons, introns, and 10 SNPs used in current analysis. Also shown is the linkage disequilibrium for each pair of markers using D’ and R2 calculated from estimated haplotype frequencies.
Table 2.
Characteristics of subjects from the Framingham Offspring Cohort
| Characteristics | Males ≤ 60 years n=391 | Males > 60 years n=477 | All Males n=868 | Premenopausal Women‡ n=126 | Postmenopausal Women n=802 | All Women n=929 |
|---|---|---|---|---|---|---|
| Age (years) | 54 ± 5 | 69 ± 6 | 62 ± 9 | 48.6 ± 4 | 62 ± 8 | 61 ± 9 |
| Weight (pounds) | 200 ± 36 | 189 ± 32 | 194 ± 34 | 156 ± 30 | 158 ± 33 | 158 ± 33 |
| Height (inches) | 69.5 ± 2.6 | 68.3 ± 2.6 | 68.9 ± 2.7 | 64.7 ± 2.4 | 63.3 ± 2.4 | 63.5 ± 2.5 |
| Physical Activity Scale† | 186 ± 87 | 122 ± 69 | 151 ± 84 | 169 ±76 | 128 ±69 | 133 ±71 |
| Currently smoking | 65 (17%) | 27 (17%) | 92 (11%) | 17 (13%) | 111 (14%) | 128 (14%) |
| Estrogen use (current/never) (%)§ | 316 (39%) | 316 (34%) |
Values are mean ± SD, unless otherwise specified.
As examined using the Physical Activity Scale for Elderly (PASE) questionnaire, which produces indices of physical activity without units.
Postmenopausal women were defined by complete cessation of menses since more than 1 year or cessation of menses for less than 1 year but taking estrogen replacement
Past users of estrogen replacement therapy (ERT) represented only 0.6% of all women, and were included among never users in the following analyses.
Linear regression analyses revealed that 6 out of the 10 SNPs in LRP5 were associated with one or more of the femur or spine BMDs in men or women after adjustment for covariates, and these associations differed between genders. In men ≤age 60 years, 3 SNPs were most significantly associated with BMD: SNP #6 in Exon 10 with femoral neck BMD (p=0.01) and Ward’s BMD (See Figure 2); SNP #5 in Exon 9 with Ward’s BMD (p=0.02); and SNP #10 with trochanter BMD (p=0.03). In men ≤ 60 years, the increase to R2 by adding these individual SNPs to the models containing covariates ranged from 1.4–1.6%. There were no significant associations in men over age 60 years.
Figure 2.
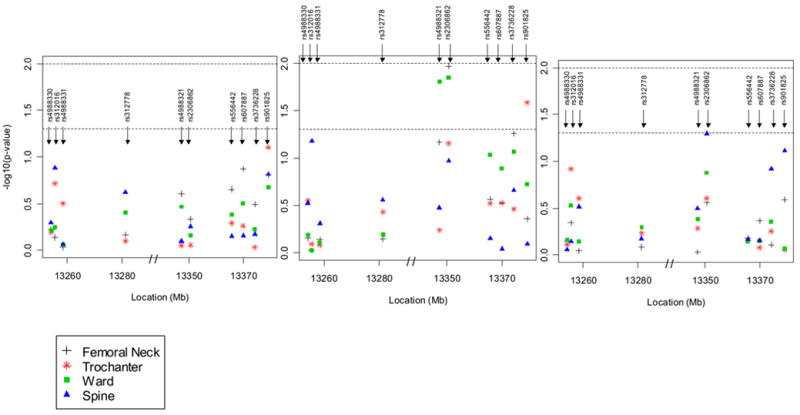
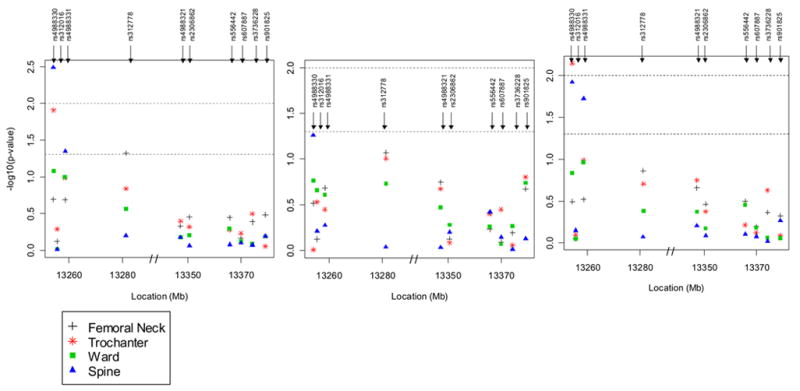
a Multivariate adjusted (age, age2, weight, physical activity) p-values for association between 10 SNPs in the LRP5 gene and BMD at the hip (femoral neck, trochanter, Ward’s area) and spine in men. The y-axis provides the –log10 of the p-value. The first of the three panels is for all men combined, the second, for men ≤ 60 years and the third, for men > 60 years. The lower dotted horizontal line is placed at a p-value of 0.05 and the upper dotted line, at a p-value of 0.01.
b Multivariate adjusted (age, age2, weight, physical activity, and estrogen use) p-values for association between 10 SNPs in the LRP5 gene and BMD at the hip (femoral neck, trochanter, Ward’s area) and spine in women. The first of the three panels is for all women combined, the second, for premenopausal women and the third, for postmenopausal women. The lower dotted horizontal line is placed at a p-value of 0.05 and the upper dotted line, at a p-value of 0.01.
In women, 3 SNPs, all situated in intron 2, were significantly associated with BMD (Figure 2): SNP #1 with trochanter BMD (p=0.01) and spine BMD (p=0.003) in all women, and trochanter BMD (p=0.007) and spine BMD (p=0.01) in postmenopausal females; SNP #4 with femoral neck BMD in all women (p=0.05); and SNP #3 with spine BMD in all women (p=0.04) and in postmenopausal women (p=0.02). In women, the increase to R2 by adding these individual SNPs to the models containing covariates ranged from 0.4–0.8%. Although each of the significant SNPs only contributed a small amount to the variation in any of the BMD measures, when all ten SNPs were simultaneously entered into the regression model adjusting for all covariates, the SNPs explained between 5.4% (for the femoral neck) and 7.4% (for the spine) of the total BMD variation in men ≤60 years and as much as 2.6% of the trochanter BMD and 2.7% of spine BMD in postmenopausal women. The number of premenopausal women was too small to calculate the R2 in this group (n=58).
To estimate the effect-size of these significant SNPs on BMD, we calculated the percent difference in adjusted BMDs according to the number of rare alleles (0, 1, or 2). In general for most of the SNPs that were significantly associated with BMD, each additional rare allele (SNP #1, SNP #3 and SNP #4) accounted for 2–4% differences in BMD among women, and for 3–5% BMD difference among men (SNP #5, SNP #6 and SNP #10). Figure 3 displays the consistency of two of the representative associations between SNP #6 in men 60 years of age and younger, and SNP #1 in women, and adjusted hip and spine BMDs. Haplotype analysis confirmed genotypes associations, without improving upon the information from the individual SNPs (data not shown).
Figure 3.
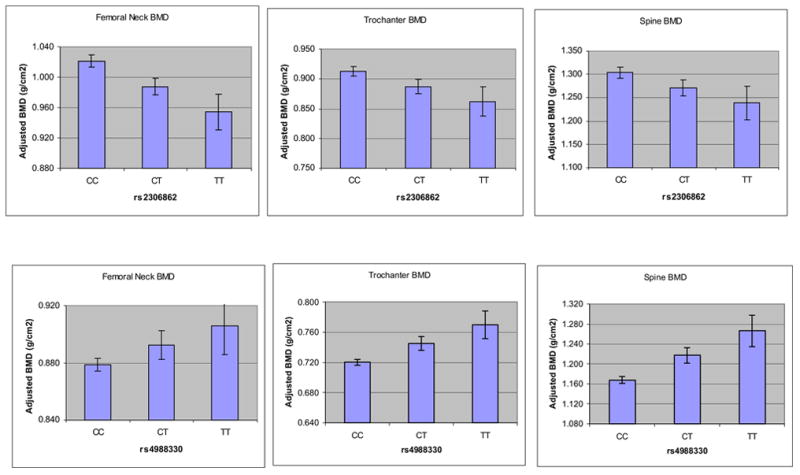
Association between two SNPs (rs2306862 in men ≤ 60 years and rs4988330 in women) in the LRP5 gene and multivariate adjusted BMD (least squares means ± SE) at the femoral neck, trochanter and spine. All associations were significant (p<0.05) except for rs2306862 in men at the spine (p=0.11).
We then specifically focused on our primary hypothesis of an interaction between genotype and physical activity. In all men, there was a significant interaction between SNP #6 in exon 10 (p-value for interaction = 0.02), and a borderline significant interaction between SNP #9 in exon 18 and physical activity on BMD of the spine (p-values for interaction = 0.05) despite the fact that the main genotype effects of these two SNPs were observed for the hip in men ≤ 60 years. Hence, the CC homozygotes demonstrated a positive association between physical activity and spine BMD, whereas for the TT homozygotes, PASE scores were inversely associated with BMD, and the CT heterozygotes showed no correlation (See Figures 4a and 4b). Despite the fact that the main genotype effects of these two SNPs were observed for the hip in men ≤ 60 years, these interactions were specific for the spine as they were not observed at other skeletal sites in men. Moreover, no interaction between PASE and any LRP5 allele was observed in females (all p-values ≥ 0.15).
Figure 4.
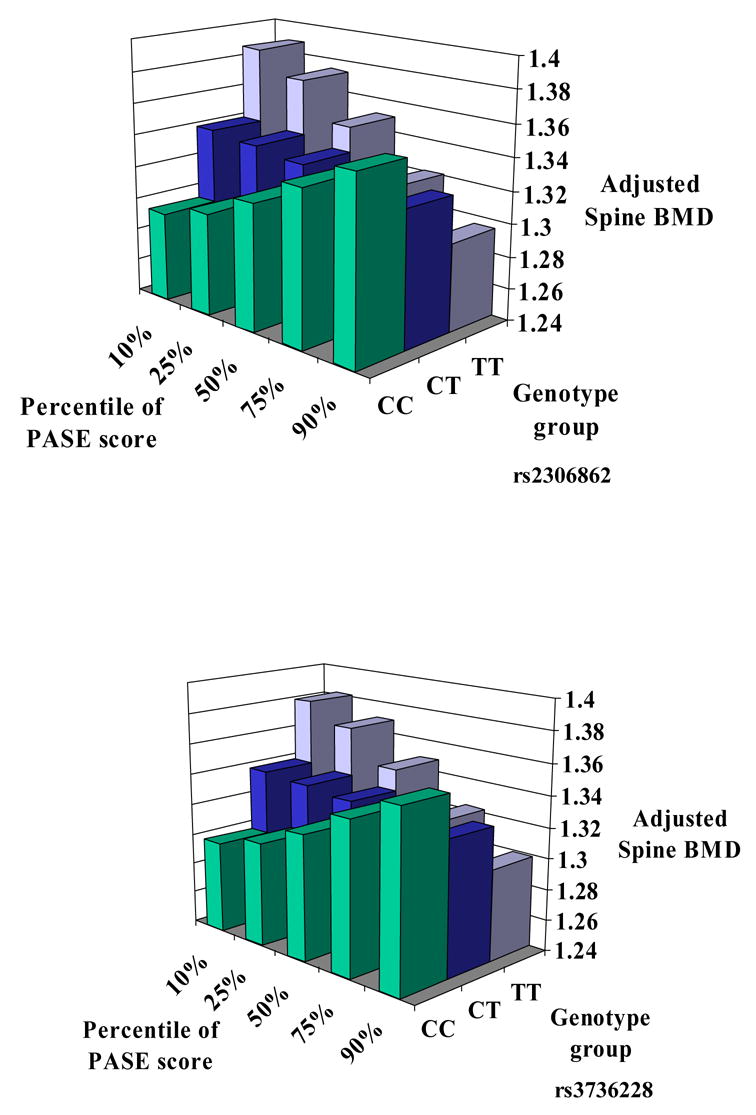
Significant interaction between two SNPs (rs2306862 and rs3736228) and physical activity (measured by PASE) on BMD of the spine in men. The x-axis provides continuously measured PASE scores divided into percentiles, the y-axis provides adjusted BMD of the lumbar spine, and the z-axis provides the three genotype groups for each SNP. For rs2306862 there were 535 men with CC, 192 men with CT and 18 men with TT genotypes. For rs3736228 there were 529 men with CC, 187 men with CT and 16 men with TT genotypes.
To evaluate the functional significance of LRP5 T or C alleles in exon 10 and 18, we expressed in HEK293 cells three common haplotypes encompassing SNP #5, #6, #7, and #9 in exon 9, 10, 15 and 18, namely G-C-A-C (haplotype 0, wild-type), G-T-G-T (haplotype 1) and A-T-G-T (haplotype 2) and measured TCF-Lef reporter activity in response to canonical Wnt3a signaling (figure 5). LRP5 haplotypes 1 and 2, which carry the T allele of exon 10 and 18, both had significantly reduced Wnt3a-stimulated TCF-Lef activity compared to wild-type alleles. Co-expressing MesD, a chaperone molecule which facilitates translocation of LRP5 to the cell membrane, increased Wnt-stimulated activity of LRP5 haplotype 0 (3.5 fold greater than control vs 2.1 fold without MesD) and haplotype 2 (1.5 fold greater than control vs 1.1 fold without MesD). Yet, in the presence of MesD, Wnt-stimulated luciferase activity remained significantly lower in cells expressing heplotypes 1 and 2 compared to wild-type (Figure 5). Next, cells expressing wild-type LRP5 and MesD were co-transfected with increasing concentrations (0.1 to 100 ng cDNA/well) of Dkk1 construct, an LRP5 antagonist. At the maximal concentration, Dkk1 completely abolished Wnt-stimulated luciferase activity (data not shown), whereas at 2–5 ng Dkk1 cDNA/well, some luciferase activity remained detectable. In these conditions, the TCF-Lef response to Wnt was similar among LRP5 haplotypes 0, 1 and 2 led to similar results (data not shown). These observations suggest that LRP5 alleles, particularly SNP #9 in exon 18 (V1330A variant), decrease Wnt signaling independent of Dkk1.
Figure 5.
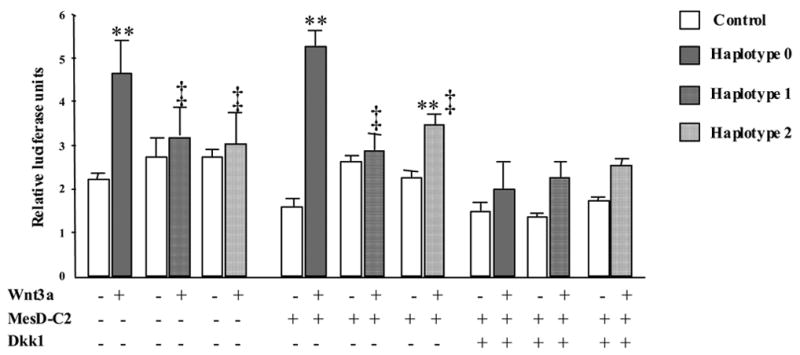
Functional differences between LRP5 haplotype 0 (wild type, G-C-A-C, closed bars), 1 (G-T-G-T, hatched bar) and 2 (A-T-G-T, dotted bar) were evaluated in HEK293 cells with or without co-expression vectors for Wnt3a, MesD and Dkk1. Canonical signaling response was measured by the TCF-Lef (Topflash®) reporter assay and expressed as luciferase units relative to an internal control (Renilla luciferase). Bars represent mean ± SE of triplicates from one experiment out of three independent experiments with similar results. **, p≤0.01 vs control (no Wnt, open bar); ‡, p<0.05 vs haplotype 0.
DISCUSSION
In this large association study in middle and older age Caucasian men and women from a relatively homogeneous population, living in Framingham, MA, we detected significant (p<0.05) evidence of association with both hip and spine BMD using multiple SNP tags and missense substitutions in the LRP5 gene. Although individual SNPs accounted for a small proportion of the variation (<2%) in BMD of the hip and spine, when all of the SNPs were considered simultaneously, a substantial proportion of variance in spine BMD was explained by genetic variation at the LRP5 locus (as much as 7.4% in men ≤ 60 years, and as much as 2.7% in post-menopausal women). Furthermore, for polymorphisms significantly associated with BMD, the difference in adjusted BMDs according to the number of rare alleles ranged between 2–5%. Not only did SNPs in LRP5 explain a greater proportion of BMD variance in younger men than women, but also the SNPs associated with BMD in men and women were grouped in different genomic regions. It is possible therefore that SNPs in the more 5′-region of LRP5 could interact with X-linked genes and/or influence transcriptional regulation of LRP5 by gonadal steroids in women. Alternatively, intronic tag SNPs upstream of the LRP5 coding and promoter regions could be markers for genetic variation in other genes on 11q12–13 contributing to BMD variance in females.46, 47 Finally, our finding that SNPs in exons 10 and 18 modified the influence of physical activity on spine BMD in men represents the first demonstration that the LRP5 gene may be implicated in the adaptation of bone to mechanical load in humans, as previously suggested in mice.26, 48 Thus, LRP5 genetic variation and, in particular, its interaction with physical activity might explain some gender-related differences in bone mass.49
There have been several previous studies of variations in the LRP5 gene and BMD. Although all have used different study designs and types of populations,27–29, 50 several SNPs found to be associated with BMD in these studies are concordant with our own observations. An association study between BMD of the radius and nine candidate genes in 481 healthy Japanese women aged 54.2±12.4 years50 reported significant associations with BMD at radius for three SNPs in LRP5, including SNP #6 in exon10 and SNP #9 in exon 18. A case-control study of 126 women with osteoporosis and 131 normal control women by the same investigators confirmed an association between two of the LRP5 SNPs and osteoporosis, although the ages of the women, and other potential confounding factors were not considered.50
Ferrari and colleagues investigated 5 common LRP5 polymorphisms in 889 male and female children and adults in Switzerland. They identified a missense substitution, SNP #5 and haplotypes of SNP #5 with SNP #9 that were significantly associated with bone mineral content and projected bone area and BMD of the lumbar spine in adult men. Altogether, LRP5 polymorphisms accounted for as much as 10–15% of the trait variance in men, but no significant associations were found for women.28 A third study confirmed both linkage and association of LRP5 alleles with BMD in 152 osteoporotic probands, their families and 160 women with BMD greater than 2.5 standard deviations above young normals, all recruited from a single region in England.29 In keeping with our own findings that LRP5 SNPs #5 and #6 in exons 9 and 10 were associated with BMD in men, but not women, the latter study reported within-family association between an intronic SNP near exon 21 and BMD of the hip, which was stronger in males. Moreover, linkage analysis revealed linkage between SNP #9 and total hip and femoral neck BMD, and comparing osteoporotic probands to unrelated postmenopausal women with high BMD revealed three other SNPs in exons 8 (rs545382), 9 (rs2277268), and 21 (IVS21 +52c<a) to be associated with BMD. Finally, Koller and colleagues confirmed an association of several LRP5 polymorphisms, including SNP #9, with BMD at the hip and spine in 1301 premenopausal women taking part in a sibling pair study using both population-based and family based association approaches.27 Despite the large number of observations, associations in these younger women were somewhat weaker compared to the other studies, supporting the hypothesis of a potentially stronger effect of LRP5 genotypes in men and older women.18
Noteworthy, one of the most commonly identified LRP5 SNPs for BMD in these studies was the SNP #9. In a large cohort from the Netherlands, this SNP has very recently been identified to be more strongly associated with projected bone area of the spine and fragility fractures in men than in women30. Our results now indicate that this polymorphism as well as SNP #6, that is in strong LD with SNP #9 (Figure 2), interact with physical activity in men, suggesting that SNP association with bone mass at the spine could result from a differential response of the axial skeleton to mechanical loading. In the Gothenburg Osteoporosis and Obesity Determinants (GOOD) study, trabecular volumetric BMD was associated with the level of overall physical activity in young men, whereas cortical volumetric BMD was not.51 Our own observation of a unique association between spine BMD and PASE score in men is fully consistent with these findings. Based on the observation that CC homozygotes for SNP#9 showed a positive association between PASE and spine BMD, whereas heterozygotes showed no correlations and TT homozygotes showed an inverse relation (albeit the latter was a small group), we propose that allelic differences in LRP5 may alter the bone remodeling balance in response to physical exercise. Thus, substitution of valine to alanine at position 1330 (i.e. C>T) could alter LRP5-mediated Wnt signaling such that catabolic signals provided by physical exercise would predominate over anabolic signals.
In keeping with this hypothesis, modulation by genetic variants of the skeletal response induced by strenuous exercise has previously been reported 16, 17. By expressing exon 18 alleles in the context of their haplotype in vitro, we found the T variant to be associated with a significantly decreased response to canonical Wnt3a signaling. Since this inhibition was observed independently of the co-expression of MesD chaperone molecule and the LRP5 antagonist Dkk1, we suggest that functional alterations associated with the LRP5 1330V variant do not necessarily involve trafficking to the cell membrane nor receptor interaction with Dkk1. Using a similar in vitro assay, Crabbe and coll. recently reported that S356L and S455L mutations in exon 6 of LRP5 identified in some men with idiopathic osteoporosis also inhibited Wnt signal transduction.52 It appears therefore that amino acid changes in various propeller modules of the LRP5 extracellular domain have the potential to influence bone mass acquisition, particularly in males. Additional experiments will be required to elucidate the actual mechanisms for altered Wnt signaling with LRP5 variants and, particularly, their osteoblastic response to mechanical stress.
Our study had several strengths including the sample size, SNP selection across the LRP gene high enough heterozygosity to enable us to maximize our power to detect association, and use of a well-validated physical activity self-report instrument (PASE) 37,39 to examine an interaction between physical activity and LRP5 polymorphisms on BMD in men specifically. It is possible therefore that differences in muscle strength or physical activity between men and women could modulate the influence of LRP5 polymorphisms on bone mass that in some previous studies have demonstrated gender-specific effects. 28–30
There were several potential limitations to our study. First there were some inconsistencies between the significant main effects of a SNP #6 mostly on the hip in men ≤60 years and the interaction of this SNP with physical activity in all men on spine BMD. Second, in all association studies there are concerns that results with p<0.05 may be false positives. Spurious allelic associations may result from population stratification; however, such risks may have been overestimated.53 Another cause of false positives arises from multiple comparisons, which we acknowledge in our analyses. A large meta-analysis recently suggested that there was replication and evidence of a true association in around one in three initial positive association studies of common variants.54 Thus, given the size of our study, the absence of population stratification in the Framingham Cohort55 and the replication of findings from other groups, it is unlikely that our findings are false positives. Finally, since data from the International Hap Map Project were not available at the time this work was done, we selected our SNPs based on the published literature at the time. Nevertheless, we examined our SNP locations in the context of the new Hap Map data (http://www.hapmap.org). Three of our SNPs (SNP#1, SNP#5 and SNP#10) were not genotyped as part of the Hap Map consortium; however our SNPs spanned the whole gene.
In conclusion, this population-based association study confirms work from previous studies that natural variation in and around LRP5 does influence variability in bone mineral density, with a suggestion of a stronger association in relatively younger adult males. The influence of any one polymorphism was small, but taken together, the proportion of variance explained by all studied polymorphisms was not inconsequential, and BMD differences conferred by the number of rare alleles were of a magnitude to be considered clinically significant, particularly in men. Perhaps the most provocative findings in our study were that LRP5 variants influence the effects of physical activity on spine bone density in men and Wnt signaling in vitro. Future studies in human populations are needed to further substantiate these findings of the interaction with regard to fracture risk in later years.
Acknowledgments
Supported by a grant from the National Institutes of Health (R01 AR/AG 41398). From the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. This work was supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nguyen TV, Blangero J, Eisman JA. Genetic epidemiological approaches to the search for osteoporosis genes. J Bone Miner Res. 2000;15(3):392–401. doi: 10.1359/jbmr.2000.15.3.392. [DOI] [PubMed] [Google Scholar]
- 2.Zmuda JM, Cauley JA, Ferrell RE. Recent progress in understanding the genetic susceptibility to osteoporosis. Genet Epidemiol. 1999;16(4):356–67. doi: 10.1002/(SICI)1098-2272(1999)16:4<356::AID-GEPI3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 3.Seeman E. Pathogenesis of bone fragility in women and men. Lancet. 2002;359(9320):1841–50. doi: 10.1016/S0140-6736(02)08706-8. [DOI] [PubMed] [Google Scholar]
- 4.Orwoll ES, Belknap JK, Klein RF. Gender specificity in the genetic determinants of peak bone mass. J Bone Miner Res. 2001;16(11):1962–71. doi: 10.1359/jbmr.2001.16.11.1962. [DOI] [PubMed] [Google Scholar]
- 5.Karasik D, Cupples LA, Hannan MT, Kiel DP. Age, gender, and body mass effects on quantitative trait loci for bone mineral density: the Framingham Study. Bone. 2003;33(3):308–16. doi: 10.1016/s8756-3282(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari S, Rizzoli R, Bonjour JP. Genetic aspects of osteoporosis. Curr Opin Rheumatol. 1999;11(4):294–300. doi: 10.1097/00002281-199907000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23(3):303–26. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 8.Thakkinstian A, D’Este C, Eisman J, Nguyen T, Attia J. Meta-analysis of molecular association studies: vitamin D receptor gene polymorphisms and BMD as a case study. J Bone Miner Res. 2004;19(3):419–28. doi: 10.1359/JBMR.0301265. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Ralston SH, Bennett ST, Brandi ML, Grinberg D, Karassa FB, Langdahl B, van Meurs JB, Mosekilde L, Scollen S, et al. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. Jama. 2004;292(17):2105–14. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 10.Mann V, Hobson EE, Li B, Stewart TL, Grant SF, Robins SP, Aspden RM, Ralston SH. A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest. 2001;107(7):899–907. doi: 10.1172/JCI10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Pottelbergh I, Goemaere S, Kaufman JM. Bioavailable estradiol and an aromatase gene polymorphism are determinants of bone mineral density changes in men over 70 years of age. J Clin Endocrinol Metab. 2003;88(7):3075–81. doi: 10.1210/jc.2002-021691. [DOI] [PubMed] [Google Scholar]
- 12.Gennari L, Brandi ML. Genetics of male osteoporosis. Calcif Tissue Int. 2001;69(4):200–4. doi: 10.1007/s00223-001-1049-3. [DOI] [PubMed] [Google Scholar]
- 13.Brown LB, Streeten EA, Shuldiner AR, Almasy LA, Peyser PA, Mitchell BD. Assessment of sex-specific genetic and environmental effects on bone mineral density. Genet Epidemiol. 2004;27(2):153–61. doi: 10.1002/gepi.20009. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari S. Genetics, Nutrition and Bone Health. In: Holick MF, Dawson-Hughes B, editors. Nutrition and Bone Health. Nutrition and Bone Health: The Humana Press Inc; 2004. pp. 19–41. [Google Scholar]
- 15.McGuigan FE, Murray L, Gallagher A, Davey-Smith G, Neville CE, Van’t Hof R, Boreham C, Ralston SH. Genetic and environmental determinants of peak bone mass in young men and women. J Bone Miner Res. 2002;17(7):1273–9. doi: 10.1359/jbmr.2002.17.7.1273. [DOI] [PubMed] [Google Scholar]
- 16.Dhamrait SS, James L, Brull DJ, Myerson S, Hawe E, Pennell DJ, World M, Humphries SE, Haddad F, Montgomery HE. Cortical bone resorption during exercise is interleukin-6 genotype-dependent. Eur J Appl Physiol. 2003;89(1):21–5. doi: 10.1007/s00421-002-0750-x. [DOI] [PubMed] [Google Scholar]
- 17.Folland J, Leach B, Little T, Hawker K, Myerson S, Montgomery H, Jones D. Angiotensin-converting enzyme genotype affects the response of human skeletal muscle to functional overload. Exp Physiol. 2000;85(5):575–9. [PubMed] [Google Scholar]
- 18.Ferrari SL, Deutsch S, Antonarakis SE. Pathogenic mutations and polymorphisms in the lipoprotein receptor-related protein 5 reveal a new biological pathway for the control of bone mass. Curr Opin Lipidol. 2005;16(2):207–14. doi: 10.1097/01.mol.0000162326.62419.e4. [DOI] [PubMed] [Google Scholar]
- 19.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–23. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 20.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70(1):11–9. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346(20):1513–21. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 22.Van Wesenbeeck L, Cleiren E, Gram J, Beals RK, Benichou O, Scopelliti D, Key L, Renton T, Bartels C, Gong Y, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet. 2003;72(3):763–71. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson ML, Gong G, Kimberling W, Recker SM, Kimmel DB, Recker RB. Linkage of a gene causing high bone mass to human chromosome 11 (11q12-13) Am J Hum Genet. 1997;60(6):1326–32. doi: 10.1086/515470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157(2):303–14. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babij P, Zhao W, Small C, Kharode Y, Yaworsky PJ, Bouxsein ML, Reddy PS, Bodine PV, Robinson JA, Bhat B, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18(6):960–74. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 26.Johnson ML, Harnish K, Nusse R, Van Hul W. LRP5 and Wnt signaling: a union made for bone. J Bone Miner Res. 2004;19(11):1749–57. doi: 10.1359/JBMR.040816. [DOI] [PubMed] [Google Scholar]
- 27.Koller DL, Ichikawa S, Johnson ML, Lai D, Xuei X, Edenberg HJ, Conneally PM, Hui SL, Johnston CC, Peacock M, et al. Contribution of the LRP5 Gene to Normal Variation in Peak BMD in Women. J Bone Miner Res. 2005;20(1):75–80. doi: 10.1359/JBMR.041019. [DOI] [PubMed] [Google Scholar]
- 28.Ferrari SL, Deutsch S, Choudhury U, Chevalley T, Bonjour JP, Dermitzakis ET, Rizzoli R, Antonarakis SE. Polymorphisms in the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with variation in vertebral bone mass, vertebral bone size, and stature in whites. Am J Hum Genet. 2004;74(5):866–75. doi: 10.1086/420771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koay MA, Woon PY, Zhang Y, Miles LJ, Duncan EL, Ralston SH, Compston JE, Cooper C, Keen R, Langdahl BL, et al. Influence of LRP5 polymorphisms on normal variation in BMD. J Bone Miner Res. 2004;19(10):1619–27. doi: 10.1359/JBMR.040704. [DOI] [PubMed] [Google Scholar]
- 30.van Meurs JB, Rivadeneira F, Jhamai M, Hugens W, Hofman A, van Leeuwen JP, Pols HA, Uitterlinden AG. Common genetic variation of the low-density lipoprotein receptor-related protein 5 and 6 genes determines fracture risk in elderly white men. J Bone Miner Res. 2006;21(1):141–50. doi: 10.1359/JBMR.050904. [DOI] [PubMed] [Google Scholar]
- 31.Ferrari SL, Deutsch S, Baudoin C, Cohen-Solal M, Ostertag A, Antonarakis SE, Rizzoli R, de Vernejoul MC. LRP5 gene polymorphisms and idiopathic osteoporosis in men. Bone. 2005;37(6):770–5. doi: 10.1016/j.bone.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Ferrari SL, Karasik D, Liu J, Karamohamed S, Herbert AG, Cupples LA, Kiel DP. Interactions of interleukin-6 promoter polymorphisms with dietary and lifestyle factors and their association with bone mass in men and women from the Framingham Osteoporosis Study. J Bone Miner Res. 2004;19(4):552–9. doi: 10.1359/JBMR.040103. [DOI] [PubMed] [Google Scholar]
- 33.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110(3):281–90. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 34.Karasik D, Myers RH, Cupples LA, Hannan MT, Gagnon DR, Herbert A, Kiel DP. Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham Study. J Bone Miner Res. 2002;17(9):1718–27. doi: 10.1359/jbmr.2002.17.9.1718. [DOI] [PubMed] [Google Scholar]
- 35.Hannan MT, Felson DT, Dawson-Hughes B, Tucker KL, Cupples LA, Wilson PW, Kiel DP. Risk factors for longitudinal bone loss in elderly men and women: the Framingham Osteoporosis Study. J Bone Miner Res. 2000;15(4):710–20. doi: 10.1359/jbmr.2000.15.4.710. [DOI] [PubMed] [Google Scholar]
- 36.Washburn RA, Smith KW, Jette AM, Janney CA. The Physical Activity Scale for the Elderly (PASE): development and evaluation. J Clin Epidemiol. 1993;46(2):153–62. doi: 10.1016/0895-4356(93)90053-4. [DOI] [PubMed] [Google Scholar]
- 37.Washburn RA, McAuley E, Katula J, Mihalko SL, Boileau RA. The physical activity scale for the elderly (PASE): evidence for validity. J Clin Epidemiol. 1999;52(7):643–51. doi: 10.1016/s0895-4356(99)00049-9. [DOI] [PubMed] [Google Scholar]
- 38.Washburn RA, Ficker JL. Physical Activity Scale for the Elderly (PASE): the relationship with activity measured by a portable accelerometer. J Sports Med Phys Fitness. 1999;39(4):336–40. [PubMed] [Google Scholar]
- 39.Schuit AJ, Schouten EG, Westerterp KR, Saris WH. Validity of the Physical Activity Scale for the Elderly (PASE): according to energy expenditure assessed by the doubly labeled water method. J Clin Epidemiol. 1997;50(5):541–6. doi: 10.1016/s0895-4356(97)00010-3. [DOI] [PubMed] [Google Scholar]
- 40.Tucker KL, Chen H, Hannan MT, Cupples LA, Wilson PW, Felson D, Kiel DP. Bone mineral density and dietary patterns in older adults: the Framingham Osteoporosis Study. Am J Clin Nutr. 2002;76(1):245–52. doi: 10.1093/ajcn/76.1.245. [DOI] [PubMed] [Google Scholar]
- 41.Zee RY, Myers RH, Hannan MT, Wilson PW, Ordovas JM, Schaefer EJ, Lindpaintner K, Kiel DP. Absence of linkage for bone mineral density to chromosome 12q12–14 in the region of the vitamin D receptor gene. Calcif Tissue Int. 2000;67(6):434–9. doi: 10.1007/s002230001175. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Deng M, Chen T, Waterman MS, Sun F. A dynamic programming algorithm for haplotype block partitioning. Proc Natl Acad Sci U S A. 2002;99(11):7335–9. doi: 10.1073/pnas.102186799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Twells RC, Mein CA, Phillips MS, Hess JF, Veijola R, Gilbey M, Bright M, Metzker M, Lie BA, Kingsnorth A, et al. Haplotype structure, LD blocks, and uneven recombination within the LRP5 gene. Genome Res. 2003;13(5):845–55. doi: 10.1101/gr.563703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29(2):311–22. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 45.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen H, Zhang YY, Long JR, Xu FH, Liu YZ, Xiao P, Zhao LJ, Xiong DH, Liu YJ, Dvornyk V, et al. A genome-wide linkage scan for bone mineral density in an extended sample: evidence for linkage on 11q23 and Xq27. J Med Genet. 2004;41(10):743–51. doi: 10.1136/jmg.2004.020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koller DL, Rodriguez LA, Christian JC, Slemenda CW, Econs MJ, Hui SL, Morin P, Conneally PM, Joslyn G, Curran ME, et al. Linkage of a QTL contributing to normal variation in bone mineral density to chromosome 11q12–13. J Bone Miner Res. 1998;13(12):1903–8. doi: 10.1359/jbmr.1998.13.12.1903. [DOI] [PubMed] [Google Scholar]
- 48.Akhter MP, Wells DJ, Short SJ, Cullen DM, Johnson ML, Haynatzki GR, Babij P, Allen KM, Yaworsky PJ, Bex F, et al. Bone biomechanical properties in LRP5 mutant mice. Bone. 2004;35(1):162–9. doi: 10.1016/j.bone.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 49.Seeman E. During aging, men lose less bone than women because they gain more periosteal bone, not because they resorb less endosteal bone. Calcif Tissue Int. 2001;69(4):205–8. doi: 10.1007/s00223-001-1040-z. [DOI] [PubMed] [Google Scholar]
- 50.Mizuguchi T, Furuta I, Watanabe Y, Tsukamoto K, Tomita H, Tsujihata M, Ohta T, Kishino T, Matsumoto N, Minakami H, et al. LRP5, low-density-lipoprotein-receptor-related protein 5, is a determinant for bone mineral density. J Hum Genet. 2004;49(2):80–6. doi: 10.1007/s10038-003-0111-6. [DOI] [PubMed] [Google Scholar]
- 51.Lorentzon M, Mellstrom D, Ohlsson C. Association of amount of physical activity with cortical bone size and trabecular volumetric BMD in young adult men: the GOOD study. J Bone Miner Res. 2005;20(11):1936–43. doi: 10.1359/JBMR.050709. [DOI] [PubMed] [Google Scholar]
- 52.Crabbe P, Balemans W, Willaert A, van Pottelbergh I, Cleiren E, Coucke PJ, Ai M, Goemaere S, van Hul W, de Paepe A, et al. Missense mutations in LRP5 are not a common cause of idiopathic osteoporosis in adult men. J Bone Miner Res. 2005;20(11):1951–9. doi: 10.1359/JBMR.050705. [DOI] [PubMed] [Google Scholar]
- 53.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361(9357):598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 54.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33(2):177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 55.Wilk JB, Manning AK, Dupuis J, Cupples LA, Larson MG, Newton-Cheh C, Demissie S, DeStefano AL, Hwang SJ, Liu C, et al. No evidence of major population substructure in the Framingham Heart Study. Genetic Epidemiology. 2005;29(3):286. [Google Scholar]


