Abstract
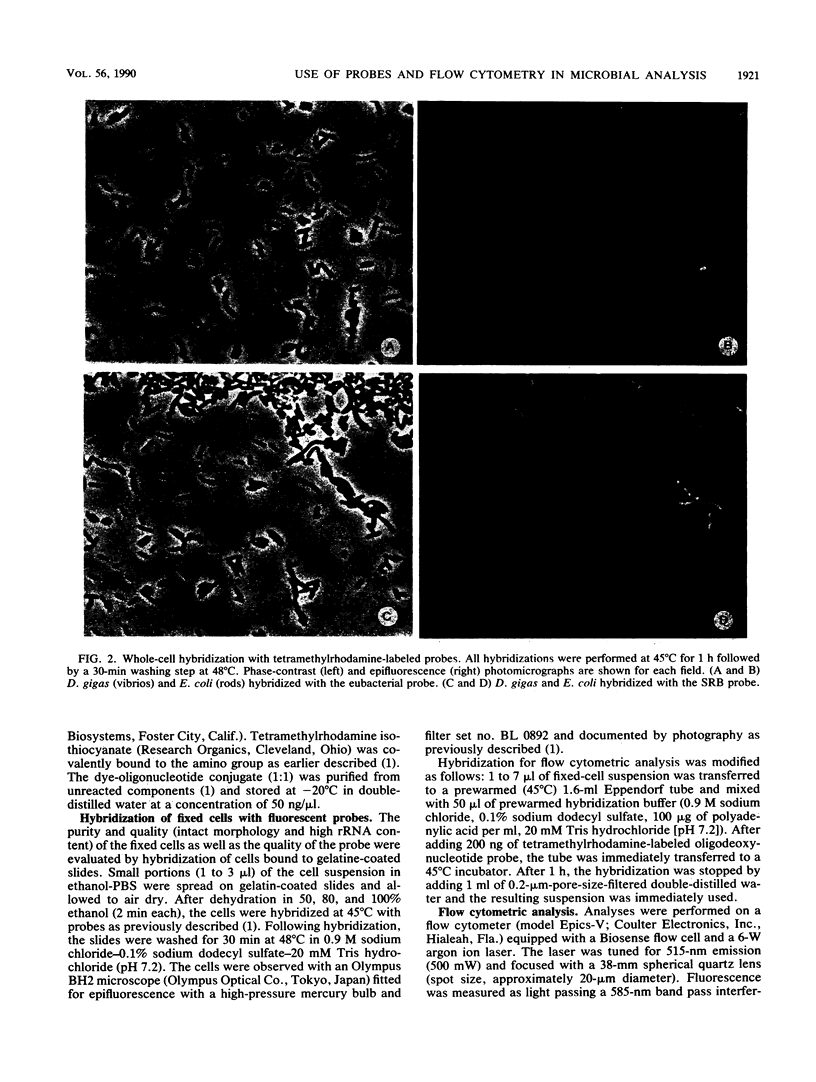
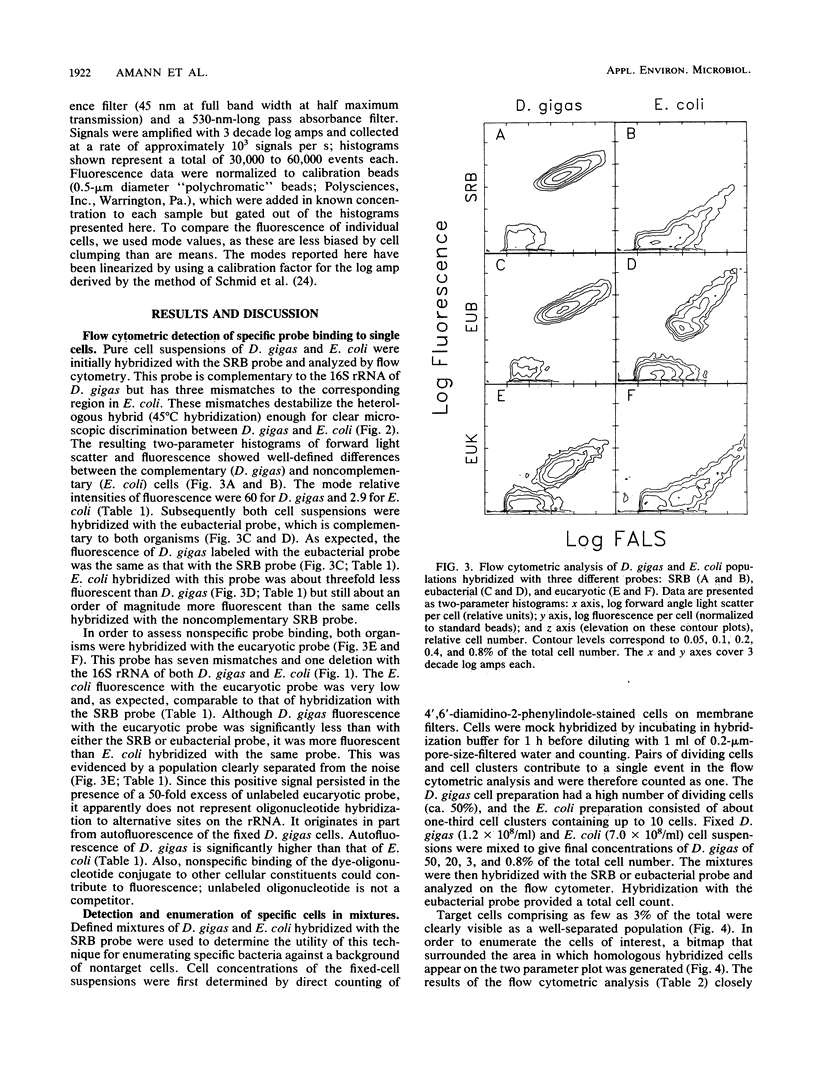
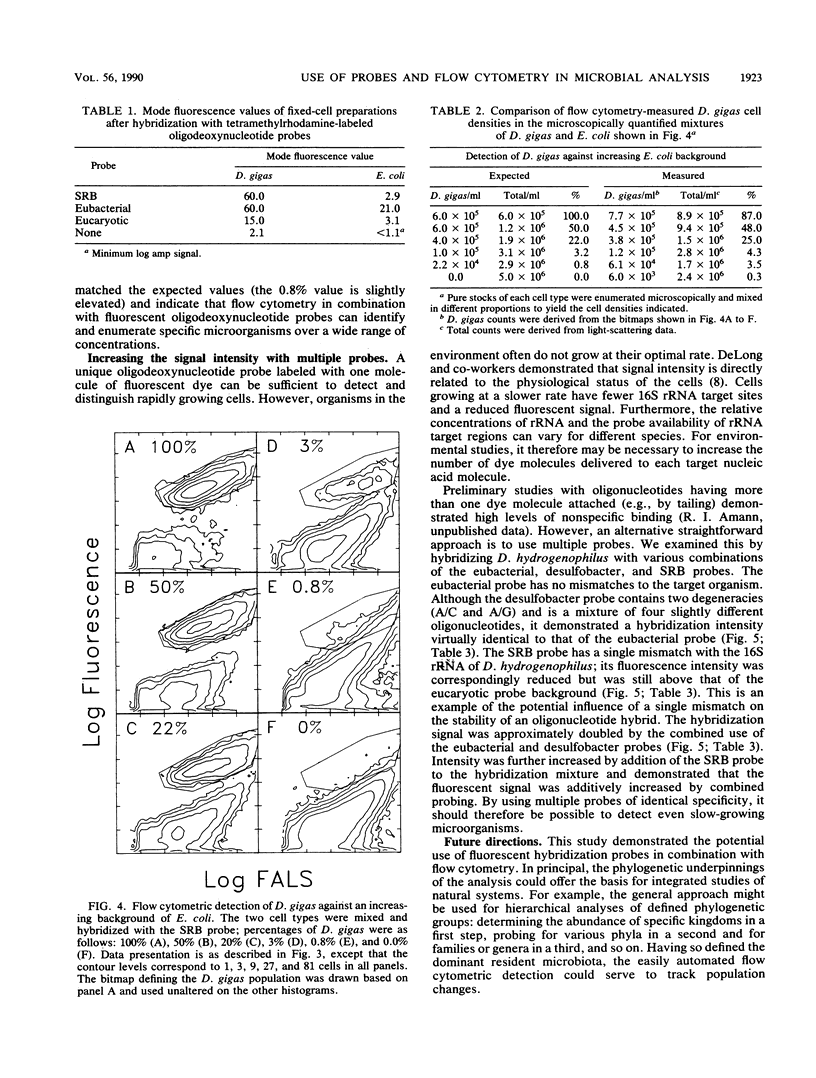
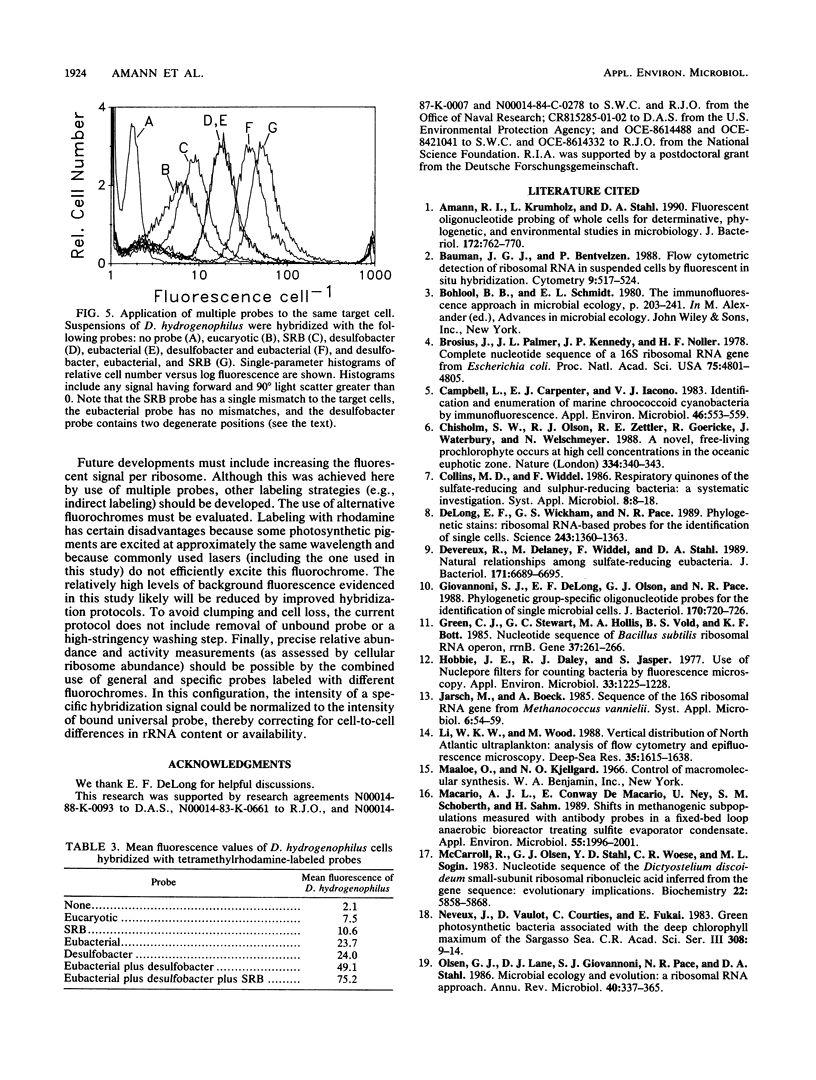
Fluorescent oligonucleotide hybridization probes were used to label bacterial cells for analysis by flow cytometry. The probes, complementary to short sequence elements within the 16S rRNA common to phylogenetically coherent assemblages of microorganisms, were labeled with tetramethylrhodamine and hybridized to suspensions of fixed cells. Flow cytometry was used to resolve individual target and nontarget bacteria (1 to 5 microns) via probe-conferred fluorescence. Target cells were quantified in an excess of nontarget cells. The intensity of fluorescence was increased additively by the combined use of two or three fluorescent probes complementary to different regions of the same 16S rRNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman J. G., Bentvelzen P. Flow cytometric detection of ribosomal RNA in suspended cells by fluorescent in situ hybridization. Cytometry. 1988 Nov;9(6):517–524. doi: 10.1002/cyto.990090602. [DOI] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L., Carpenter E. J., Iacono V. J. Identification and enumeration of marine chroococcoid cyanobacteria by immunofluorescence. Appl Environ Microbiol. 1983 Sep;46(3):553–559. doi: 10.1128/aem.46.3.553-559.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Devereux R., Delaney M., Widdel F., Stahl D. A. Natural relationships among sulfate-reducing eubacteria. J Bacteriol. 1989 Dec;171(12):6689–6695. doi: 10.1128/jb.171.12.6689-6695.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario A. J., Conway de Macario E., Ney U., Schoberth S. M., Sahm H. Shifts in methanogenic subpopulations measured with antibody probes in a fixed-bed loop anaerobic bioreactor treating sulfite evaporator condensate. Appl Environ Microbiol. 1989 Aug;55(8):1996–2001. doi: 10.1128/aem.55.8.1996-2001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Robertson B. R., Button D. K. Characterizing aquatic bacteria according to population, cell size, and apparent DNA content by flow cytometry. Cytometry. 1989 Jan;10(1):70–76. doi: 10.1002/cyto.990100112. [DOI] [PubMed] [Google Scholar]
- Schmid I., Schmid P., Giorgi J. V. Conversion of logarithmic channel numbers into relative linear fluorescence intensity. Cytometry. 1988 Nov;9(6):533–538. doi: 10.1002/cyto.990090605. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. B., Carlucci A. F. Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl Environ Microbiol. 1985 Aug;50(2):194–201. doi: 10.1128/aem.50.2.194-201.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]



