Abstract
Transglutaminases (TGases; EC 2.3.2.13) are a family of enzymes that catalyze calcium-dependent covalent cross-linking of cellular proteins by establishing ɛ-(γ-glutamyl)lysine isopeptide bonds. These covalent isopeptide bonds are of great physiological significance because they are highly resistant to proteolysis, denaturants, and reducing agents. Prior studies have demonstrated the presence of isopeptide bonds in the sheath and cuticle of filarial parasites, suggesting an important role for TGase-catalyzed reactions during the growth and development of filarial nematodes. Herein we report the identification and cloning of a cDNA encoding a TGase from the dog heartworm Dirofilaria immitis (DiTG). The DiTG expressed in Escherichia coli (recombinant DiTG) was able to catalyze calcium-dependent cross-linking reactions. The derived amino acid sequence of the DiTG cDNA (pDiTG) predicts a protein of 57.1 kDa and includes an N-terminal hydrophobic signal peptide. The pDiTG has no sequence similarity with any of the known TGases, but it has significant homology to protein disulfide isomerase (PDI) and, particularly, to the PDI-related endoplasmic reticulum protein ERp60, a PDI isoform found in the lumen of endoplasmic reticulum. As predicted from the amino acid sequence homology, recombinant DiTG catalyzed the isomerization of intramolecular disulfide/sulfhydryl bonds in denatured RNase in vitro as effectively as did mammalian PDI. Conversely, purified PDI from bovine liver could catalyze protein cross-linking reactions in a Ca2+-dependent manner. This report describes the dual catalytic activity of TGase and PDI in post- and/or cotranslational modification of newly synthesized proteins. These TGase-catalyzed posttranslational modifications may play a pivotal role in the synthesis of new cuticle during the growth and maturation of filarial parasites.
Filarial nematodes that cause chronic infections in human and animal populations are responsible for considerable morbidity in their hosts and thus pose a major health problem in many parts of the world (1). Although there are effective prophylactic agents that can prevent infection by larval stages, at present there is no safe and reliable chemotherapeutic agent that is active against adult worms of filarial species. This problem is further compounded by the acquired resistance to conventional insecticides seen in vector populations (1). Therefore, identification of key enzymes and molecules that are essential for the growth and survival of nematodes may offer targets for developing effective chemotherapeutic agents and vaccines.
The outer surface of all nematodes consists of a multilayered cuticle, a complex structure that serves as an exoskeleton, interacts with the host’s immune system, and functions as an absorptive surface (2, 3). The entire cuticle is shed at each molt and replaced with a new cuticle synthesized by the underlying layer of hypodermal tissue, a large syncytium that extends throughout the length of the nematode (3). Both the synthesis and secretion of cuticular components by the hypodermis are tightly coupled to the molting cycles (2). The structure of the cuticle can vary widely between species and between developmental stages within a species. Despite this diversity, the basic components of cuticle include (i) collagen-like proteins that form the medial and basal layers of the cuticle, (ii) noncollagenous proteins that form the epicuticular and external cortical regions, and (iii) nonstructural proteins associated with the external surface. The collagens that are cross-linked by disulfide bonds can be solubilized only with buffers containing detergents and 2-mercaptoethanol (3, 4). The noncollagenous proteins, referred to as cuticlins, exhibit unusual chemical properties and are proteins cross-linked by both reducible and nonreducible bonds. The nonreducible interchain covalent bonds are of two types, ɛ-(γ-glutamyl)lysine isopeptide bonds and bonds between tyrosine side chains (5). Prior studies have reported the presence of ɛ-(γ-glutamyl)lysine isopeptide bonds catalyzed by transglutaminase (TGase) in the developing embryos and microfilarial sheath of Litomosoides carinii and Brugia malayi and in the infective larvae of Onchocerca volvulus (6, 7).
This laboratory has demonstrated the presence of the protein cross-linking enzyme TGase and TGase-catalyzed products in the human filarial parasite B. malayi (6, 8). More recently, enzymatically active TGases (pTGase) with approximate molecular masses of 56 kDa were purified from adult worms of B. malayi and Dirofilaria immitis species (9, 10). Biochemical studies suggested that pTGases from these two parasites are very similar but that their properties are distinct from the mammalian TGases (9, 10). In this report, we describe the molecular cloning and characterization of a cDNA encoding an enzymatically active TGase from D. immitis. We also report that, in addition to its cross-linking activity, recombinant D. immitis TGase (rDiTG) was able to refold a denatured RNase into its active form, an activity characteristic of protein disulfide isomerase (PDI) and PDI-related proteins.
MATERIALS AND METHODS
Parasites and Parasite Antigens.
D. immitis parasites used in this study were originally derived from a single dog obtained through the U.S.–Japan Cooperative Medical Sciences Program, National Institutes of Health. Infection of mosquitoes and collection of 0-hr L3 (mosquito-derived infective stage larvae), 48-hr L3 (48-hr after in vitro culture), and 6-day L4 (6 days after in vitro culture of 0-hr L3) were carried out as described (11). D. immitis soluble antigens from adult worms and total antigens from larvae and larval excretory-secretory (E-S) products were prepared essentially as described (12, 13). For production of adult D. immitis E-S products, worms were incubated in NCTC135/Iscove’s modified Dulbecco’s medium (GIBCO/BRL). Culture supernatant fractions containing E-S products were collected and passed through 0.45-μm (pore size) filters, concentrated by using Centriplus-10 concentrators (Millipore), and dialyzed against PBS. Protein concentrations were measured relative to a BSA standard by using the bicinchoninic acid microassay reagent as described in the manufacturer’s protocol (Pierce).
PAGE.
Soluble and E-S antigens of D. immitis were subjected to SDS/PAGE in 4–20% gradient gels (NOVEX, San Diego) under reducing conditions. After electrophoresis, proteins were transferred to nitrocellulose (Optitran, Schleicher & Schuell). Blots were developed with rabbit antibodies raised against a synthetic peptide corresponding to the N-terminal sequence of B. malayi pTGase (10) or rabbit anti-rDiTG antiserum (see below). Bound antibody was detected with alkaline phosphatase-conjugated goat anti-rabbit IgG and the substrates nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (GIBCO/BRL). Two-dimensional nonequilibrium pH-gradient gel electrophoresis was performed by using ampholines from pH 5.0 to 8.0 (Pharmacia) in the first dimension (14) and then a 4–20% SDS/PAGE gel in the second dimension.
Amino Acid Sequencing.
Native 56-kDa TGase protein from D. immitis adult female parasite extracts (10) was separated by two-dimensional SDS/PAGE and then transferred to a poly(vinylidene difluoride) membrane, and the spot corresponding to D. immitis TGase was excised from the membranes. Seventeen such spots were pooled and used for N-terminal sequence analysis in an automated protein sequencer (Applied Biosystems). For internal amino acid sequence analysis, spots containing DiTG were excised from preparative two-dimensional SDS/PAGE gels of adult female worm extracts stained with Coomassie brilliant blue R-250. Forty-eight such spots were pooled and subjected to a partial trypsin digestion in the gel. The digested protein sample was separated by HPLC and the peptides were sequenced as described above.
Preparation of cDNA and Larval cDNA Library.
RNA was isolated from adult worms by using TRI REAGENT (Molecular Research Center, Cincinnati) according to the supplier’s protocol. Poly(A)+ mRNA was prepared from total RNA by using the Oligotex Direct mRNA kit (Qiagen, Chatsworth, CA) by the manufacturer’s instructions. A 6-day L4 larval cDNA expression library was constructed in the Unizap XR vector (Stratagene) according to the manufacturer’s protocol. Total RNA was extracted from 140,000 L4 larvae. cDNA was synthesized from 7 μg of poly(A)+ mRNA and cloned unidirectionally, and the library was packaged by using Stratagene Gigapack II packaging extracts according to the manufacturer’s instructions. The titer of the amplified library was 1.05 × 109 plaque-forming units/ml, 98% of the clones being recombinants.
Cloning, Sequencing, Expression, and Purification of DiTG.
First-strand cDNA synthesized from poly(A)+ RNA was used as a template for PCRs. Degenerate primers P1 and P2 were synthesized corresponding to the N-terminal amino acid sequence (MKFTDADFKEG; sense) and to a tryptic peptide (MFVVYGK, antisense). PCR conditions consisted of an initial cycle of denaturation at 95°C for 3 min followed by 35 additional cycles (95°C for 15 s, 57°C for 15 s, and 72°C for 1 min). A D. immitis L4 larval cDNA library was screened by DNA hybridization as described (15).
To obtain the 5′ region of the message, first-strand cDNA was amplified by using the nematode splice leader sequence (SL1) (16) as a sense primer (P3, 5′-GGTTTAATTACCCAAGTTTGAG-3′) and a primer derived from the initial 707-nucleotide PCR product as an antisense primer (P4, 5′-TCCCTCCTTGAAGTCCGCATCTGTAAATTTCAT-3′) (see Results).
The nucleotide sequence of the gene encoding DiTG was obtained by the dideoxynucleotide chain-termination method by using an Applied Biosystems 373A DNA sequencer. The sequence information was compiled and analyzed by using macvector′ (Eastman Kodak), assemblylign′ (Eastman Kodak), and pc/gene (Intelligenitics) software packages. A search for homologous sequences was conducted in a nonredundant protein sequence database through the National Center for Biotechnology Information, by using the BLAST network.
The entire coding sequence of DiTG except the signal sequence was subcloned into a plasmid expression vector, pTrcHisB (Invitrogen). The plasmid was transfected into Escherichia coli, and the protein expression was monitored by SDS/PAGE and immunoblot analysis by using a T7 Tag antibody (Novagen). The recombinant protein (rDiTG) was purified by using the Talon metal affinity resin (CLONTECH) under nondenaturing conditions as in the manufacturer’s protocol.
Production of Polyclonal Antibodies to rDiTG.
A rabbit was immunized subcutaneously with 75 μg of the purified rDiTG protein in complete Freund’s adjuvant (Sigma), followed by three subsequent immunizations of the same dose of rDiTG mixed in incomplete Freund’s adjuvant (Sigma). Bleeding and immunization were performed in alternate weeks. Sera were separated and stored at −70°C until use. The IgG-enriched fraction from anti-rDiTG antiserum was obtained by 50% ammonium sulfate precipitation, followed by extensive dialysis against 0.1 M sodium phosphate/0.15 M NaCl, pH 7.2 (PBS). The Ig content was determined by measuring absorbance at OD280 and comparing the absorbance with that of a blank PBS control.
Enzyme Assays.
TGase activity was determined in a microtiter plate assay as described (9). One milliunit of TGase activity was defined as the change in A490 min−1 generated by 1 ng of purified guinea pig liver TGase (Sigma). The PDI activity was determined essentially as described (17). One unit of PDI activity is defined as that catalyzing reactivation of 1 unit of RNase per min; 1 RNase unit is defined as the amount producing a change in A260 of 1 absorbance unit/min (17). Negative controls for both assays included BSA (Sigma) and an unrelated recombinant protein expressed in pTrcHisB.
RESULTS
Protein Sequence of Native DiTG.
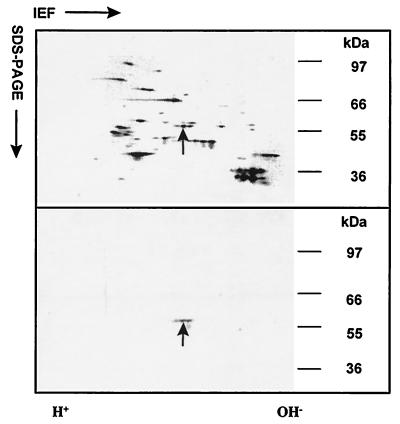
Antibody raised against a synthetic peptide representing the N-terminal 12 amino acids of B. malayi pTGase (10) recognized a 56-kDa protein in immunoblots of crude extracts of D. immitis adult female worms. In immunoblots of D. immitis adult female extracts subjected to two-dimensional SDS/PAGE, the antiserum identified two immunoreactive protein spots at 56 kDa (Fig. 1). Both the protein spots were sequenced and found to have identical N-terminal sequences, and one of the spots was used for determining the internal amino acid sequence. The sequence of the N-terminal 29 amino acids of the immunoreactive D. immitis protein contained 12 residues that were identical to the N-terminal amino acid sequence of B. malayi pTGase (data not shown). Three tryptic peptides obtained as a result of partial digestion of the 56-kDa TGase protein were sequenced, and their positions within the deduced amino acid sequence encoded by the DiTG cDNA are shown in Fig. 2.
Figure 1.
Two-dimensional nonequilibrium pH-gradient gel electrophoresis–SDS/PAGE separation of protein extracts from D. immitis adult female worm. Silver-stained gel (Upper) and two-dimensional immunoblot (Lower) probed with rabbit antibodies to B. malayi pTGase show the 56-kDa protein spots (arrow). The two immunoreactive spots (Lower) had identical N-terminal sequences, and one of the spots (arrow) was used to determine the internal amino acid sequence of the protein.
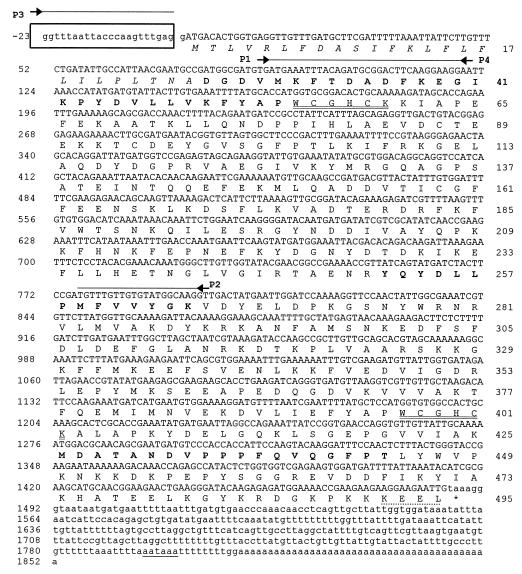
Figure 2.
Nucleotide and predicted amino acid sequences of the DiTG cDNA. The splice leader (SL1) 22-nucleotide sequence is boxed. The nucleotide sequence is numbered on the left, and the deduced amino acid sequence is numbered on the right. The consensus polyadenylylation signal is underlined. Lowercase type on the nucleotide line represents untranslated regions. Primers used for PCR amplifications are indicated by arrows above the nucleotide line. The predicted signal peptide residues are shown in italic type. The thioredoxin/PDI active sites are double underlined and the endoplasmic reticulum retention sequence (KEEL) is indicated by a dotted line. The amino acid sequences obtained from the native protein are shown in boldface type.
Molecular Cloning of D. immitis TGase.
A 707-bp product was obtained by PCR amplification using the first-strand cDNA and two degenerate primers (P1 and P2) deduced from D. immitis pTGase peptide sequences. The 707-bp PCR-amplified product had an ORF encoding an amino acid sequence identical to N-terminal sequence derived from the native TGase protein and extending beyond the primer P1 sequence (Fig. 2). The PCR-derived 707-nucleotide cDNA was then used as a probe for screening a D. immitis larval cDNA library and two hybridizing clones were identified. The longest of the two the clones (1,472 nucleotides) was sequenced and found to have the complete 3′ end of the cDNA including a translational stop codon (TAA), a polyadenylylation signal (AATAAA), and a 42-bp poly(A)+ tail (Fig. 2). However, this clone lacked about 300 nucleotides of the sequence contained in the 5′ end of the 707-nucleotide PCR product that was used to screen the cDNA library. The missing 5′ end of the cDNA was obtained by PCR using the SL1 primer (P3) and a sequence-specific antisense primer (P4). This yielded a fragment of 143 nucleotides containing the SL1 sequence (Fig. 2), an ORF of 120 nucleotides, and a potential 22-amino acid signal peptide preceding the N terminus of the mature protein.
Characterization of DiTG cDNA.
The composite cDNA sequence encoding DiTG is 1,875 nucleotides long and encodes a single ORF of 1,485 bases. The ORF codes for a protein of 495 amino acids, including a signal peptide of 25 residues (Fig. 2). The N-terminal and internal amino acid sequences obtained from the purified protein were identified in the amino acid sequence deduced from the DiTG cDNA. The calculated Mr of 54,353 for the mature protein is in agreement with the 56-kDa native protein observed on SDS/PAGE (Fig. 1).
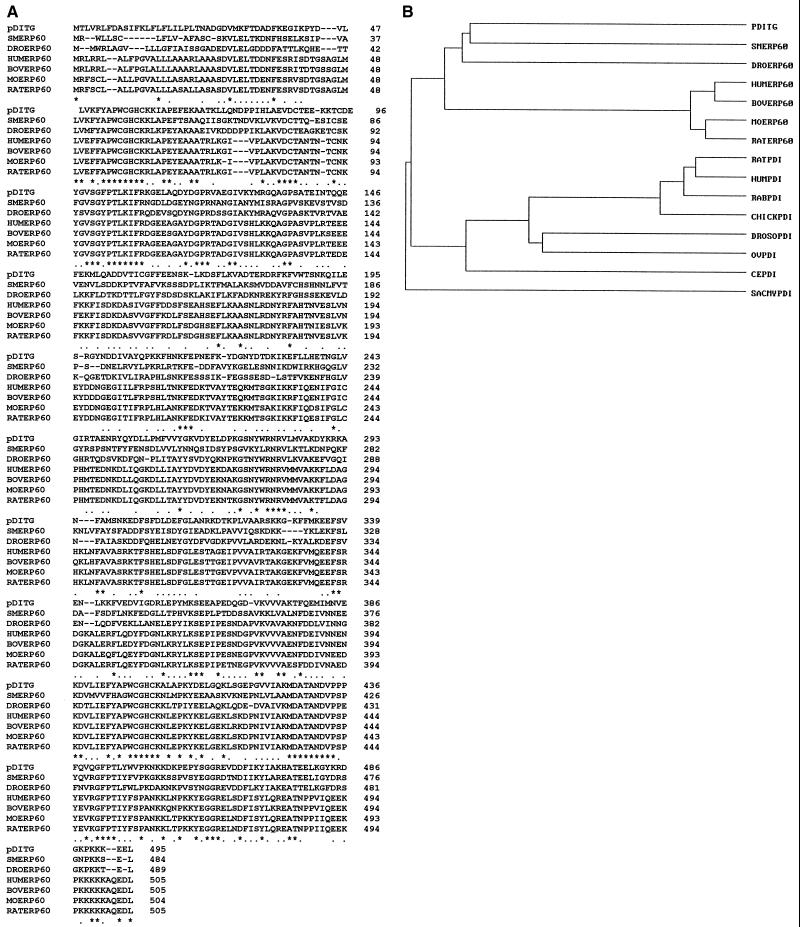
Interestingly, derived amino acid sequence of the DiTG cDNA (pDiTG) revealed no homology with any of the known TGases in the GenBank database. However, it contained two distinct regions at residues 55–60 and 397–402 that are identical to the putative active-site sequences of the ERp60/PDI/thioredoxin family of proteins (Fig. 2). A multiple sequence alignment (18) of pDiTG with ERp60 from several species showed 43–52% identity relative to pDiTG (Fig. 3A). In addition, pDiTG had 31–35% identity with PDIs from various organisms (data not shown). A dendrogram built from the pair-wise similarity scores using the average linkage cluster analysis (19) shows a closer relationship of pDiTG to ERp60 than to PDI (Fig. 3B). The pDiTG sequence includes a C-terminal endoplasmic reticulum retention sequence (KEEL) (20), suggesting that, like PDI and ERp60, DiTG is a luminal endoplasmic reticulum protein.
Figure 3.
(A) Comparison of pDiTG sequence with ERp60 from Schistosoma mansoni (SMERp60, GenBank accession no. P38658), Drosophila melanogaster (DROERp60, locus 1699220), human (HUMERp60, U75885), bovine (BOVERp60, P38657), mouse (MOERp60, P27773), and rat (RATERp60, P11598) by using the multiple (clustal) alignment (17) that gives the highest number of matched residues. Identical residues are marked with an asterisk, and conserved residues are marked with a dot. Gaps, marked by hyphens, have been introduced for better alignment. (B) A dendrogram showing the relationship of pDiTG with ERp60 and PDI proteins from various species. PDI proteins compared include those from rat (GenBank accession no. P54001), human (HUMPDI, P55059), rabbit (RABPDI, P21195), chicken (CHICKPDI, P16924), D. melanogaster (DROSOPDI, P54399), Onchocerca volvulus (OVPDI, U12440), Caenorhabditis elegans (CEPDI, Q10576), and Saccharomyces cerevisiae (SACMYPDI, locus 68468).
Protein Expression and Enzyme Activity of rDiTG.
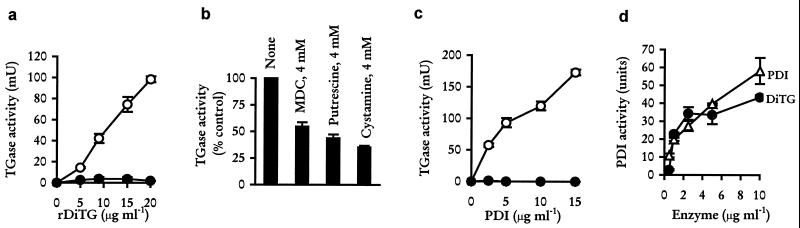
The cDNA corresponding to the mature protein was subcloned into a plasmid expression vector, pTrcHisB, and expressed as an N-terminal histidine fusion protein (rDiTG). rDiTG was purified by metal-chelating chromatography under nondenaturing conditions and tested for TGase activity by using a microtiter plate assay (9). rDiTG catalyzed a time-dependent (data not shown) and dose-dependent increase in the incorporation of a primary amine substrate, biotinylated pentylamine (BPA), into dimethylated casein (Fig. 4a). Addition of EDTA (Fig. 4a) or EGTA (data not shown) to the reaction mixture completely abrogated the enzyme activity, suggesting that, like with other known TGases, the reaction was calcium-dependent. rDiTG-catalyzed conjugation of BPA was shown to be specific, as evidenced by the significant inhibition of the reaction by various structurally unrelated enzyme-specific inhibitors including monodansylcadaverine, putrescine, and cystamine (Fig. 4b). Because the deduced amino acid sequence of DiTG cDNA shared significant sequence homology with mammalian PDI, the ability of mammalian PDI to function as a TGase enzyme was evaluated. Indeed, PDI from bovine liver (Sigma) could effectively catalyze calcium-dependent incorporation of BPA into dimethylated casein (Fig. 4c). In addition, rDiTG was able to catalyze the isomerization of disulfide bond interchange as demonstrated by its ability to reactivate a denatured RNase (Fig. 4d). The presence of Ca2+ was not essential for either PDI- or rDiTG-catalyzed disulfide isomerization reactions because the presence of EDTA in the reaction mixture did not affect the isomerase activity.
Figure 4.
TGase and disulfide isomerase activities associated with rDiTG and purified bovine liver PDI proteins. The ability of rDiTG (a and b) and bovine liver PDI (c) to conjugate biotinylated pentylamine to dimethylated casein was determined in the presence of either Ca2+ alone (○), Ca2+ plus EDTA (•), or Ca2+ plus the indicated TGase inhibitors (b). Also, the ability of purified bovine PDI and rDiTG to renature denatured RNase in the presence of 2.5 mM EDTA was determined (d). Each data point represents the mean ± SEM of triplicate values from two experiments.
Identification of Native Antigen Corresponding to rDiTG.
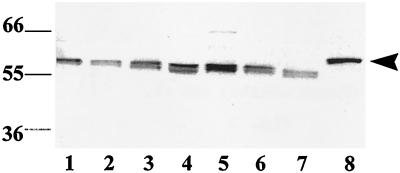
Serum from a rabbit immunized with rDiTG was tested for reactivity to native antigens in D. immitis parasite extracts prepared from 0-hr L3, 48-hr L3, 6-day L4, and male and female adult worms and E-S products from larvae and adults. IgG-enriched anti-rDiTG antisera immunoreacted to a 56-kDa native antigen in parasite extracts from all stages and in larval and adult E-S products (Fig. 5). Serum from a preimmune rabbit did not react with any antigens in the parasite extracts (data not shown). These data suggest that TGase is ubiquitously expressed in all developmental stages of D. immitis and is also released by larvae and adults in culture in vitro. It was interesting to note the presence of TGase in the E-S products despite the observation that pDiTG has an endoplasmic reticulum-retention signal (20). Prior studies have shown the presence of endoplasmic reticulum-resident proteins in sites other than endoplasmic reticulum (21). However, the significance of the presence of DiTG in E-S products is not known.
Figure 5.
Identification of native DiTG in the developmental stages of D. immitis. Immunoblot analysis was carried out with IgG-enriched anti-rDiTG antiserum and the following parasite extracts. Lanes: 1, 0-hr L3 (200); 2, 48-hr L3 (200); 3, 6-day L4 (200); 4, larval E-S products (10 μg); 5, adult female worm (10 μg); 6, adult male worm (10 μg); 7, adult E-S products (10 μg); 8, rDiTG (100 ng,  ).
).
DISCUSSION
Antibodies raised against a synthetic peptide corresponding to the B. malayi TGase protein cross-reacted with DiTG (10). Consequently, this property was exploited to purify the cross-reactive protein from D. immitis by two-dimensional SDS/PAGE (Fig. 1) and obtain the N-terminal and internal amino acid sequences of native DiTG. The sequence of the N-terminal 29 amino acids of the D. immitis protein contained 12 residues that were identical to the N-terminal amino acid sequence of B. malayi TGase protein (data not shown). These initial results suggested that both parasites have very similar TGase proteins. This conclusion is further supported by recent studies showing that TGase enzymes from both B. malayi and D. immitis are biochemically very similar (9, 10).
The DiTG cDNA sequence reported herein was compiled from three overlapping sequences: a 707-bp PCR product, a 1,472-bp cDNA clone, and a 143-bp splice leader-PCR product. Cloning of nematode genes is greatly facilitated if they are naturally transspliced at the 5′ end with the conserved 22-nucleotide nematode splice leader SL1. Most, but not all, nematode mRNAs have the SL sequence at their 5′ ends, and the presence of the 5′ SL1 sequence is indicative of an apparent full-length cDNA (16). Therefore, in cloning the 5′ terminus of the message, we used a primer specific for the SL1 splice leader and an internal clone-specific primer. This yielded a 143-bp PCR product containing the SL1 sequence at the 5′ end, thus confirming the presence of a full-length message. Although the composite DiTG cDNA sequence was compiled based on one cDNA clone and two PCR-amplified fragments, the extensive overlap at the nucleotide and amino acid level supports the contiguity of the sequence. By using a primer specific to the 5′ end of the DiTG cDNA immediately after the leader sequence and a 3′ end primer, we amplified the full-length ORF coding for the mature protein from first-strand DNA. Sequence analysis of the resulting 1,410-bp PCR product confirmed the accuracy of the composite sequence. This 1,410-bp PCR product was expressed in a plasmid expression vector for use in enzymatic studies.
Interestingly, despite the cross-linking activity of rDiTG, the deduced amino acid sequence of DiTG has no homology to any of the TGases in the database. However, the sequence contains two distinct regions identical to the putative active site sequences of the ERp60/PDI/thioredoxin family of proteins (Fig. 3) (22). Thioredoxin serves as an essential cofactor in various oxidation–reduction reactions by inducing reversible oxidation of an active center disulfide bond (23). A number of eukaryotic proteins contain similar active-site regions and are evolutionarily related to thioredoxin (24). Of these, PDI and the PDI-related proteins (ERp60, ERp72), which contain two and three thioredoxin-family active sites, respectively, are of special interest (22). PDI is a major luminal protein in secretory cells and, like TGase, catalyzes the post- and/or cotranslational modification of newly synthesized proteins. PDI catalyzes thiol–disulfide interchange reactions leading to disulfide bond formation, isomerization, or reduction of substrate proteins (22). Multiple sequence alignment and dendrogram analysis clearly shows that pDiTG is more closely related to ERp60 than to PDI. However, recently several groups have demonstrated the PDI activity of ERp60 and ERp72 (25, 26). Therefore, because of the homology of DiTG to the PDI family of proteins, we tested rDiTG for PDI activity. Indeed, rDiTG was able to catalyze the refolding of denatured RNase to its active form (Fig. 4d). Conversely, mammalian PDI exhibited TGase activity.
The observation that mammalian PDI can function as a cross-linking enzyme is also extremely interesting. Evidence is accumulating that PDI may associate with other proteins and regulate the formation of complexes by various proteins (27). For example, in myeloid cells of chronic myelogenous leukemia patients, increased levels of PDI have been reported to regulate the formation of complexes between nuclear proteins and the regulatory domains of interferon-inducible genes (28). Recently, rat PDI was shown to promote cross-linking of smaller aggregates of substrate proteins, a behavior that has been termed anti-chaperone activity (29). By using lysozyme and alcohol dehydrogenase as substrates, it was shown that rat PDI promotes the formation of inactive disulfide protein cross-links in vitro (29, 30) and this activity requires Ca2+.
The lumen of the endoplasmic reticulum contains abundant soluble proteins (22), and from the present study it is evident that DiTG may be one among them. It is tempting to speculate that TGase may require PDI activity to promote catalysis of protein cross-linking reactions. It is possible that DiTG catalyzes the cross-linking of nascent protein chains in the endoplasmic reticulum and also helps the newly synthesized protein to fold correctly before they are transported out of endoplasmic reticulum. The observation that TGase, but not PDI, activity requires Ca2+ indicates that the active sites for these two functions may be distinct.
In conclusion, we have cloned a gene from a nematode parasite that encodes a protein with significant similarity to ERp60. The expressed recombinant protein has both TGase and PDI activities. The presence of two ERp60/thioredoxin/PDI active-site sequences in the DiTG cDNA-encoded protein, and the ability of rDiTG to renature denatured RNase, identify this ERp60-like molecule as a member of this growing multifunctional protein family. Thus, the cloning and expression of an active DiTG multifunctional enzyme provide insights that should delineate further the molecular mechanisms involved in cuticle biosynthesis in nematodes.
Acknowledgments
We thank Glenn R. Frank, Liang Tang, Carol Talkington Verser, Lauren McNitt, and Dan Stinchcomb for critical reading of the manuscript; and Jude Richard for editorial review of the manuscript. We also thank Cindy Bozic and Stephanie Berger of the Sequencing Core Group for DNA and protein sequencing. This work was supported in part by a grant to K.M. from the United Nations Development Program/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TGase, transglutaminase; E-S, excretory-secretory; DiTG, D. immitis TGase; pDiTG, amino acid sequence encoded by DiTG cDNA; rDiTG, recombinant D. immitis TGase; PDI, protein disulfide isomerase; SL1, 22-nucleotide nematode splice leader sequence.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF008300).
References
- 1.World Health Organization Expert Committee on Filariasis. Tech. Rep. Ser. No. 702. New York: WHO; 1992. [PubMed] [Google Scholar]
- 2.Inglis W G. Proc Zool Soc Lond. 1964;143:465–502. [Google Scholar]
- 3.Maizels R M, Blaxter M L, Selkirk M E. Exp Parasitol. 1993;77:380–384. doi: 10.1006/expr.1993.1096. [DOI] [PubMed] [Google Scholar]
- 4.Selkirk M E, Nielson L, Kelly C, Partono F, Sayers G, Maizels R M. Mol Biochem Parasitol. 1989;32:229–246. doi: 10.1016/0166-6851(89)90073-x. [DOI] [PubMed] [Google Scholar]
- 5.Lustigman S. Parasitol Today. 1993;9:294–297. doi: 10.1016/0169-4758(93)90128-3. [DOI] [PubMed] [Google Scholar]
- 6.Mehta K, Rao U R, Vickery A C, Birckbichler P J. Biochem Biophys Res Commun. 1990;173:1051–1057. doi: 10.1016/s0006-291x(05)80892-7. [DOI] [PubMed] [Google Scholar]
- 7.Lustigman S, Brotman B, Huima T, Mehta K, Prince A M. Antimicrob Agents Chemother. 1995;39:1913–1919. doi: 10.1128/aac.39.9.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta K, Rao U R, Vickery A C, Fesus L. Mol Biochem Parasitol. 1992;53:1–16. doi: 10.1016/0166-6851(92)90002-2. [DOI] [PubMed] [Google Scholar]
- 9.Singh R N, Mehta K. Eur J Biochem. 1994;225:625–634. doi: 10.1111/j.1432-1033.1994.00625.x. [DOI] [PubMed] [Google Scholar]
- 10.Singh R N, Chandrashekar R, Mehta K. Int J Biochem Cell Biol. 1995;27:1285–1291. doi: 10.1016/1357-2725(95)00102-U. [DOI] [PubMed] [Google Scholar]
- 11.Frank G R, Grieve R B. J Parasitol. 1991;77:950–956. [PubMed] [Google Scholar]
- 12.Grieve R B, Frank G R, Mika-Grieve M, Culpepper J A, Mok M. J Immunol. 1992;148:2511–2515. [PubMed] [Google Scholar]
- 13.Frank G R, Tripp C T, Grieve R B. Mol Biochem Parasitol. 1996;75:231–240. doi: 10.1016/0166-6851(95)02534-0. [DOI] [PubMed] [Google Scholar]
- 14.O’Farrell P Z, Goodman H M, O’Farrell P H. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 15.Chandrashekar R, Curtis K C, Weil G J. J Infect Dis. 1995;171:1586–1592. doi: 10.1093/infdis/171.6.1586. [DOI] [PubMed] [Google Scholar]
- 16.Blaxter M, Liu L. Int J Parasitol. 1996;26:1025–1033. [PubMed] [Google Scholar]
- 17.Lambert N, Freedman R B. Biochem J. 1983;213:235–243. doi: 10.1042/bj2130235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins D G, Sharp P M. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 19.Sneath P H A, Sokal R R. Numerical Taxonomy. New York: Freeman; 1973. [Google Scholar]
- 20.Munro S, Pelham H R B. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 21.Meldolesi J, Krause K-H, Michalak M. Cell Calcium. 1996;20:83–86. doi: 10.1016/s0143-4160(96)90053-6. [DOI] [PubMed] [Google Scholar]
- 22.Freedman R B, Hirst T R, Tuite M F. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 23.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 24.Gething M-J, Sambrook J. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 25.Bourdi M, Demady D, Martin J L, Jabbour S K, Martin B M, George J W, Pohl L R. Arch Biochem Biophys. 1995;323:397–403. doi: 10.1006/abbi.1995.0060. [DOI] [PubMed] [Google Scholar]
- 26.Van P N, Rupp K, Lampen A, Soling H-D. Eur J Biochem. 1993;213:789–795. doi: 10.1111/j.1432-1033.1993.tb17821.x. [DOI] [PubMed] [Google Scholar]
- 27.Luz J M, Lennarz W J. In: Stress-Inducible Cellular Responses. Feige U, Morimoto R I, Yahara I, Polla B, editors. Basel: Birkhauser; 1996. pp. 97–117. [Google Scholar]
- 28.Johnson E. J Biol Chem. 1992;267:14412–14417. [PubMed] [Google Scholar]
- 29.Puig A, Gilbert H F. J Biol Chem. 1994;269:7764–7771. [PubMed] [Google Scholar]
- 30.Primm T P, Walker K W, Gilbert H F. J Biol Chem. 1996;271:33664–33669. doi: 10.1074/jbc.271.52.33664. [DOI] [PubMed] [Google Scholar]