Abstract
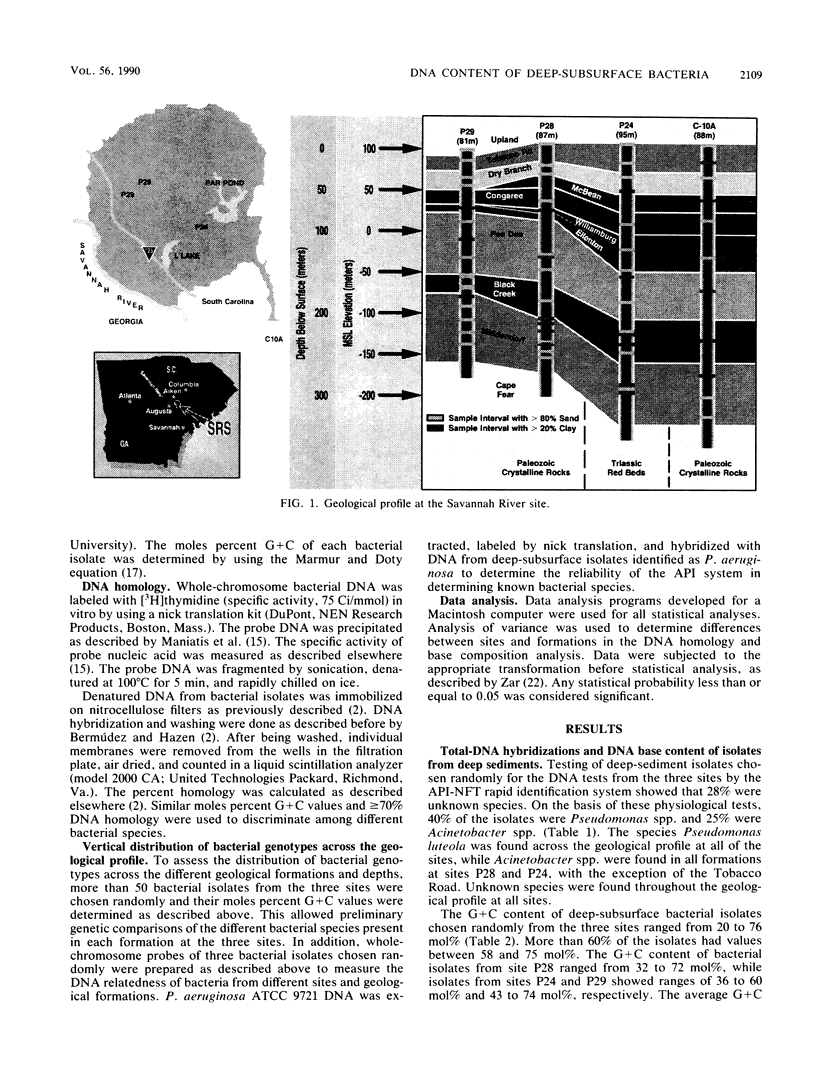
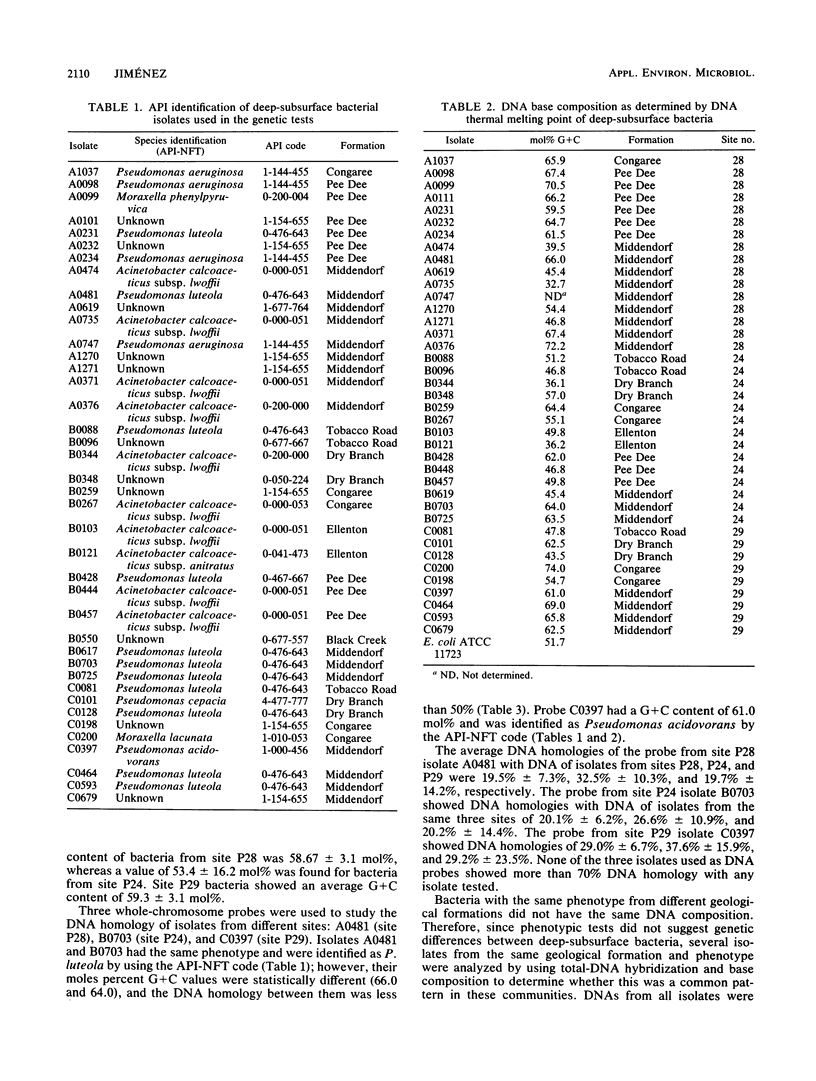
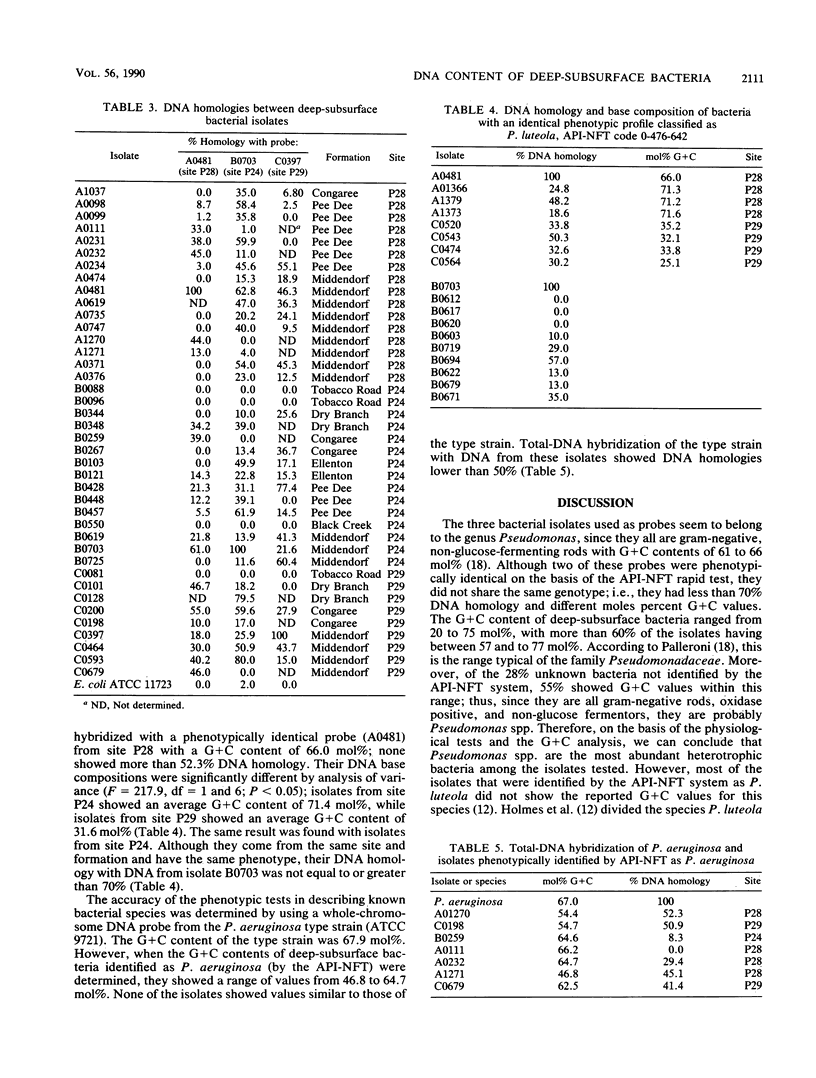
Bacterial isolates from deep-sediment samples from three sites at the Savannah River site, near Aiken, S.C., were studied to determine their microbial community composition and DNA structure by using total DNA hybridization and moles percent G + C. Standard phenotypic identification underestimated the bacterial diversity at the three sites, since isolates with the same phenotype had different DNA structures in terms of moles percent G + C and DNA homology. The G + C content of deep-subsurface bacteria ranged from 20 to 77 mol%. More than 60% of the isolates tested had G + C values similar to those of Pseudomonas spp., and 12% had values similar to those of Acinetobacter spp. No isolates from deeper formations showed the same DNA composition as isolates from upper formations. Total-DNA hybridization and DNA base composition analysis provided a better resolution than phenotypic tests for the understanding of the diversity and structure of deep-subsurface bacterial communities. On the basis of the moles percent G + C values, deep-subsurface isolates tested seemed to belong to the families Pseudomonadaceae and Neisseriaceae, which might reflect a long period of adaptation to the environmental conditions of the deep subsurface.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balkwill D. L., Fredrickson J. K., Thomas J. M. Vertical and horizontal variations in the physiological diversity of the aerobic chemoheterotrophic bacterial microflora in deep southeast coastal plain subsurface sediments. Appl Environ Microbiol. 1989 May;55(5):1058–1065. doi: 10.1128/aem.55.5.1058-1065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez M., Hazen T. C. Phenotypic and genotypic comparison of Escherichia coli from pristine tropical waters. Appl Environ Microbiol. 1988 Apr;54(4):979–983. doi: 10.1128/aem.54.4.979-983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse H., El-Banna T., Auling G. Evaluation of different approaches for identification of xenobiotic- degrading pseudomonads. Appl Environ Microbiol. 1989 Jun;55(6):1578–1583. doi: 10.1128/aem.55.6.1578-1583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell F. S. Microbiological comparison of surface soil and unsaturated subsurface soil from a semiarid high desert. Appl Environ Microbiol. 1989 Sep;55(9):2420–2423. doi: 10.1128/aem.55.9.2420-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Hicks R. J., Li S. W., Brockman F. J. Plasmid incidence in bacteria from deep subsurface sediments. Appl Environ Microbiol. 1988 Dec;54(12):2916–2923. doi: 10.1128/aem.54.12.2916-2923.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen T. C., Jiménez L. Enumeration and identification of bacteria from environmental samples using nucleic acid probes. Microbiol Sci. 1988 Nov;5(11):340–343. [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Sharp R. J., Williams R. A. Properties of Thermus ruber Strains Isolated from Icelandic Hot Springs and DNA:DNA Homology of Thermus ruber and Thermus aquaticus. Appl Environ Microbiol. 1988 Aug;54(8):2049–2053. doi: 10.1128/aem.54.8.2049-2053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaji S., Rao N. S., Saisree L., Sheth V., Reddy G. S., Bhargava P. M. Isolation and identification of Pseudomonas spp. from Schirmacher Oasis, Antarctica. Appl Environ Microbiol. 1989 Mar;55(3):767–770. doi: 10.1128/aem.55.3.767-770.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]



