Abstract
We have identified a member of the VEGF family by computer-based homology searching and have designated it VEGF-D. VEGF-D is most closely related to VEGF-C by virtue of the presence of N- and C-terminal extensions that are not found in other VEGF family members. In adult human tissues, VEGF-D mRNA is most abundant in heart, lung, skeletal muscle, colon, and small intestine. Analyses of VEGF-D receptor specificity revealed that VEGF-D is a ligand for both VEGF receptors (VEGFRs) VEGFR-2 (Flk1) and VEGFR-3 (Flt4) and can activate these receptors. However, VEGF-D does not bind to VEGFR-1. Expression of a truncated derivative of VEGF-D demonstrated that the receptor-binding capacities reside in the portion of the molecule that is most closely related in primary structure to other VEGF family members and that corresponds to the mature form of VEGF-C. In addition, VEGF-D is a mitogen for endothelial cells. The structural and functional similarities between VEGF-D and VEGF-C define a subfamily of the VEGFs.
The formation of blood vessels occurs either by the in situ differentiation of endothelial cell precursors (angioblasts) and association of these cells to form primitive vessels, a process called vasculogenesis, or by growth of preexisting vessels, a process called angiogenesis (for review, see ref. 1). Vasculogenesis establishes the primary vascular plexus of the early embryo, whereas development of blood vessels during later embryogenesis and adult life occurs primarily by angiogenesis. Angiogenesis in the adult is tightly controlled; under normal circumstances it occurs almost exclusively in the female reproductive system (2). However, angiogenesis can be activated in the adult in response to tissue damage and is important in certain pathological conditions such as tumorigenesis, rheumatoid arthritis, and diabetic retinopathy (2). Once blood vessels have been established, endothelial cells undergo tissue-specific changes to generate numerous types of functionally distinct vessels as organs differentiate (3). These processes require that endothelial cells respond to a variety of extracellular signals that activate receptors responsible for growth and differentiation.
VEGF is a homodimeric glycoprotein that is mitogenic for endothelial cells and is an angiogenic factor that acts via the endothelial-specific receptor tyrosine kinases (VEGF receptors, VEGFRs) VEGFR-1 (Flt1) and VEGFR-2 (Flk1) (for review, see ref. 4). Development of blood vessels in the embryo is dependent on VEGF as the formation of vessels in mouse embryos heterozygous for a disrupted VEGF gene was aberrant and resulted in embryonic lethality (5, 6). VEGF is also a potent inducer of vascular permeability (7).
Numerous proteins closely related in primary structure to VEGF have been reported in recent years that may also play roles in vascular biology. Placenta growth factor (PlGF) is approximately 46% identical in amino acid sequence to VEGF (8), binds to VEGFR-1 (9), and can form heterodimers with VEGF (10). Although the biological function of PlGF is unknown, it can significantly potentiate the action of low concentrations of VEGF in vitro and in vivo (9). VEGF-B, which is approximately 43% identical in amino acid sequence to VEGF, is mitogenic for endothelial cells, can form heterodimers with VEGF, and may be involved in angiogenesis in muscle and heart (11). As yet, the receptors for VEGF-B are uncharacterized. Another member of the VEGF family, VEGF-C, was isolated as a ligand for the tyrosine kinase VEGFR-3 (Flt4) (12), a receptor that is expressed in endothelial cell precursors in day 8.5 mouse embryos and later in development is expressed in venous and lymphatic endothelium (13). The pattern of VEGF-C gene expression in mouse embryos suggests that VEGF-C may regulate angiogenesis of the lymphatic vasculature (14). VEGF-C is also a ligand for VEGFR-2 (12), but the functional significance of this potential interaction in vivo is unknown. The amino acid sequence of VEGF-C has a central region that is related to other members of the VEGF family and exhibits approximately 30% identity to VEGF. In addition, the VEGF-C sequence has N-terminal and C-terminal extensions that are not present in VEGF, PlGF, or VEGF-B (12, 15). The biosynthesis of VEGF-C involves proteolytic processing that gives rise to a mature secreted protein that essentially consists of the VEGF homology domain (16), i.e., the portion of the molecule that is related in primary structure to all other members of the VEGF family and that contains the cystine knot motif that is found in VEGF family members and in other growth factors (17).
We report herein the characterization of a human cDNA for a fifth member of the VEGF family, designated VEGF-D, that is most closely related to VEGF-C in primary structure. The mouse homologue of VEGF-D was recently designated as the c-fos-induced growth factor (18). We show that VEGF-D binds to and induces tyrosine phosphorylation of the endothelial cell receptors VEGFR-2 and VEGFR-3 and that, as for VEGF-C, the capacity of VEGF-D to bind to these receptors is associated with the VEGF homology domain. In addition, VEGF-D is mitogenic for endothelial cells. Our results demonstrate the existence of a subfamily of the VEGFs that has VEGF-C and VEGF-D as founding members.
MATERIALS AND METHODS
Cloning of cDNA for VEGF-D.
Computer searches for VEGF-related sequences were carried out by using the fasta search algorithm (19). The expressed sequence tag (EST) encoding the C-terminal region of VEGF-D (GenBank accession no. H24828) had been isolated by the Integrated Molecular Analysis of Genome Expression Consortium as part of the Washington University–Merck EST Project (20). The EST was obtained from the American Type Culture Collection and was used as hybridization probe for isolation of cDNA from a human lung cDNA library (Stratagene).
Northern Blot Analysis.
A 1.1-kb fragment of the human VEGF-D cDNA, containing the region encoding amino acid residues 168–354 (Fig. 1) and approximately 500 nt of the 3′ untranslated region, was used to screen human multiple-tissue Northern blots (CLONTECH) according to manufacturer’s instructions.
Figure 1.
Comparison of human VEGF-D with other members of the VEGF family. Alignment of the deduced amino acid sequences of human VEGF-D, mouse VEGF-D (18), human VEGF-C (15), human VEGF165 (21), human VEGF-B167 (11), and human PlGF-2 (22) is shown. Residues that match the sequence of human VEGF-D are boxed. The asterisks above the hVEGF-D sequence denote the eight cysteine residues that are conserved in all VEGF family members. Arrows denote positions of proteolytic cleavage that give rise to mature VEGF-C (16). The line above the hVEGF-D sequence denotes a putative signal sequence for protein secretion (23). Potential N-linked glycosylation sites in human VEGF-D are marked by brackets above the sequence. Solid circles above the hVEGF-D sequence denote cysteine residues involved in motifs that resemble those of Balbiani ring 3 protein (CX10CXCXC) (24).
Expression of VEGF-DΔNΔC in COS Cells and Purification by Affinity Chromatography.
A DNA fragment encoding the portion of the human VEGF-D polypeptide from residues 93 to 201 was inserted into the expression vector pEFBOSSFLAG immediately downstream from DNA sequence encoding the interleukin 3 (IL-3) signal sequence (25) and the FLAG octapeptide (IBI/Kodak), so that protein synthesis would give rise to a truncated secreted VEGF-D polypeptide that was tagged with the FLAG octapeptide at its N terminus. This protein was designated VEGF-DΔNΔC. VEGF-DΔNΔC was expressed in COS cells that had been transiently transfected by the DEAE-dextran method (26). The resulting conditioned cell culture medium (approximately 150 ml), collected after 7 days of incubation, was subjected to affinity chromatography using a resin to which the M2 (anti-FLAG) mAb (IBI/Kodak) had been coupled according to the manufacturer. An identical affinity purification was carried out in parallel by using conditioned culture medium from cells that had been transfected with pEFBOSSFLAG (i.e., without VEGF-D coding sequences) to generate negative control samples for bioassays. VEGF-DΔNΔC arising from affinity chromatography was analyzed by Western blot analysis using mAb M2 (IBI/Kodak) or control antibody as described by the manufacturer.
Bioassay to Monitor Binding of Ligands to the Extracellular Domain of VEGFR-2.
A derivative of the pre-B cell line Ba/F3 (27), expressing a chimeric receptor consisting of the extracellular domain of mouse VEGFR-2 and the transmembrane and cytoplasmic domains of the mouse erythropoietin receptor (EpoR) (28), was generated and designated Ba/F3-VEGFR-2-EpoR (S.A.S., unpublished results). The Ba/F3-VEGFR-2-EpoR cells were washed three times in PBS to remove all IL-3, resuspended in cell culture medium without IL-3, and distributed into 96-well microtiter plates at a concentration of 10,000 cells per well. Fractions arising from affinity purification of VEGF-DΔNΔC were then diluted into the cell culture medium. Cells expressing a chimeric receptor, consisting of the extracellular domain of the mouse endothelial cell receptor Tie2 (29) and the transmembrane and cytoplasmic domains of mouse EpoR (S.A.S. and A.S. Runting, unpublished results), were used as a nonresponding control cell line. Cells were incubated for 48 h, during which the cells in the cell culture medium alone had died, and DNA synthesis occurring in the other wells (i.e., in the presence of VEGF-DΔNΔC) was determined by addition of 1 μCi of [3H]thymidine and quantitating incorporation over a 4-h period by β-counting.
Expression of Proteins in Baculovirus and Analysis of Receptor Stimulation.
Derivatives of human VEGF-D and VEGF-C cDNA were cloned into the baculoviral vector pFASTBAC1 (GIBCO/BRL) for generation of viral stocks. The supernatants of High Five cells (Invitrogen) were harvested 48 h after infection with virus stocks, adjusted to pH 7 with NaOH, diluted with 1 vol of DMEM containing 0.2% fetal calf serum and used to stimulate NIH 3T3 cells expressing human VEGFR-3 (30) or porcine aortic endothelial cells expressing human VEGFR-2 (31). Stimulation of cells and analysis of phosphorylated receptors were carried out as described (12).
Binding Assays with Soluble VEGFR Extracellular Domains.
For binding experiments, 293T cells were transfected with plasmids encoding the soluble receptor-Ig fusion proteins VEGFR-1-Ig (E. Korpelainen, Haartman Institute, Helsinki), VEGFR-2-Ig (Y. Gunji, Haartman Institute, Helsinki), or VEGFR-3-Ig (K. Pajusola, Biotechnology Institute, Helsinki) by using the calcium phosphate method. The cells were incubated for 24 h after transfection, washed with DMEM containing 0.2% BSA, and starved for 24 h. Medium was then collected and clarified by centrifugation, and fusion proteins were precipitated by using protein A-Sepharose beads. The Sepharose beads were then incubated at room temperature for 3 h with 900 μl of metabolically 35S-labeled medium from 293EBNA cells that had been transfected with expression plasmids encoding human VEGF-DΔNΔC, human VEGF-CΔNΔC, or human VEGF165 by using the calcium phosphate method. Metabolic labeling of 293EBNA cells had been carried out essentially as described (16). The Sepharose beads were then washed twice with binding buffer [0.5% BSA/0.02% Tween 20/heparin (1 μg/ml) in PBS] at 4°C, once with PBS and boiled in Laemmli sample buffer, and proteins were then analyzed by SDS/PAGE.
Mitogenesis Assays with Endothelial Cells.
Mitogenesis assays with bovine aortic endothelial cells (BAEs) were carried out as described (32).
RESULTS
Cloning and Characterization of cDNA for Human VEGF-D.
To identify additional members of the VEGF family, computer searches of the EMBL, GenBank, and dbEST databases were carried out with the amino acid sequences of VEGF, VEGF-B, and VEGF-C. A 419-bp cDNA sequence was identified that encodes a polypeptide that was similar in amino acid sequence to the C-terminal 100 residues of VEGF-C (12, 15). This cDNA was used to isolate human lung cDNA that contained the entire ORF for a polypeptide of 354 amino acids. The polypeptide was clearly a member of the VEGF family because it contained the characteristic cystine knot motif and other conserved amino acids found within the VEGF homology domain. We designated this human protein as VEGF-D. Recently, a mouse protein designated the c-fos-induced growth factor was reported (18) that was 85% identical in amino acid sequence to human VEGF-D. It is highly likely that c-fos-induced growth factor is the mouse homologue of VEGF-D.
Alignment of the deduced amino acid sequence of human VEGF-D with those for other members of the VEGF family is shown in Fig. 1. The eight cysteine residues thought to be involved in intra- and intersubunit disulfide bonds and that are conserved in all other members of the VEGF family are also conserved in human VEGF-D. When gaps introduced into the amino acid sequences for the purposes of alignment are ignored for calculations, human VEGF-D is 48% identical to human VEGF-C, 31% identical to human VEGF165, 28% identical to human VEGF-B167, and 32% identical to human PlGF. VEGF-D contains a VEGF homology domain (residues 101–196) that is related in sequence to all other VEGF family members. In this region, the amino acid sequences of VEGF-D and VEGF-C are 61% identical. As with VEGF-C, VEGF-D has long N- and C-terminal extensions in comparison to the other family members. These extensions in VEGF-D are related in sequence to those in VEGF-C; the N-terminal extensions exhibit 25% amino acid identity and the C-terminal extensions exhibit 37% identity. Human VEGF-D has three potential N-linked glycosylation sites. The positions and sequences of two of these sites (residues 155–157 and 185–187 of human VEGF-D) are conserved in human VEGF-C (Fig. 1). The amino acid sequence of human VEGF-D has a highly hydrophobic region at the N terminus that is typical of a signal sequence for protein secretion. The C-terminal 135-amino acid region of human VEGF-D is rich in cysteine residues. Many of these cysteine residues are located such that they resemble the spacing of the repeat units (CX10CXCXC), which are found in the Balbiani ring 3 protein (Fig. 1), a major cysteine-rich protein synthesized in the larval salivary glands of the midge Chironomus tentans (24). Such repeats are also found in the C-terminal region of VEGF-C (12, 15).
Distribution of VEGF-D mRNA in Adult Human Tissues.
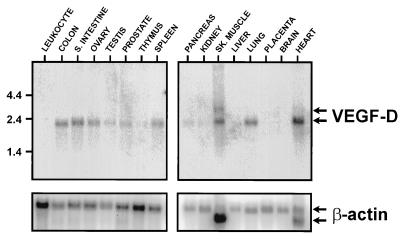
Northern blot analysis revealed that in most of the human tissues where VEGF-D mRNA was detected, the size of the transcript was 2.3 kb (Fig. 2). The only exception was skeletal muscle where an additional less-abundant transcript of 2.8 kb was detected. VEGF-D mRNA was most abundant in heart, lung, skeletal muscle, colon, and small intestine.
Figure 2.
Northern blot analyses for detection of VEGF-D mRNA in polyadenylated RNA from human tissues. (Upper) Results of hybridizations with a human VEGF-D cDNA probe. (Lower) Results with a β-actin cDNA probe after VEGF-D probe had been stripped from the filters. The sizes of RNA molecular size markers, in kb, for the VEGF-D hybridizations are shown to the left. Tissues used as sources of RNA are indicated. SK. MUSCLE, skeletal muscle; S. INTESTINE, small intestine (mucosal lining).
VEGF-D Is a Ligand for VEGFR-2.
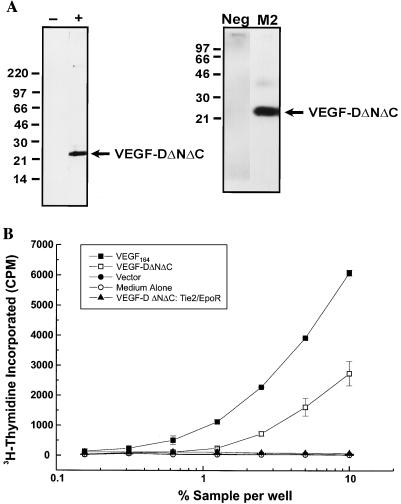
It has been shown previously that the biosynthesis of VEGF-C, the protein most closely related in primary structure to VEGF-D, involves proteolytic cleavage that gives rise to a truncated mature form of VEGF-C that binds to VEGFR-2 and VEGFR-3 (16). The mature form contains the region of VEGF-C that is homologous to all other members of the VEGF family—the VEGF homology domain. The two sites of proteolytic cleavage involved in generation of mature VEGF-C (16) are shown in Fig. 1. Our preliminary immunoprecipitation analyses of VEGF-D biosynthesis suggested that VEGF-D is processed in a similar fashion to VEGF-C (data not shown). To test whether a derivative of VEGF-D corresponding to mature VEGF-C is a ligand for VEGFR-2, a portion of human VEGF-D from amino acid residues 93 to 201 that contained the VEGF homology domain (Fig. 1) was tagged at the N terminus with the FLAG octapeptide and expressed transiently in COS cells. This FLAG-tagged derivative was designated VEGF-DΔNΔC. Subsequently, VEGF-DΔNΔC was enriched by M2 (anti-FLAG) affinity chromatography. VEGF-DΔNΔC from the enrichment was analyzed by Western blotting and silver staining (Fig. 3A), which revealed that under reducing conditions VEGF-DΔNΔC exists as a species of ∼22 kDa. This is consistent with the predicted molecular weight for this internal fragment of VEGF-D (Mr, 12,800) plus N-linked glycosylation (VEGF-DΔNΔC contains two potential N-linked glycosylation sites). Under nonreducing conditions, VEGF-DΔNΔC existed as a species of ∼25 kDa (data not shown). Cross-linking studies have subsequently demonstrated that a proportion of the VEGF-DΔNΔC secreted by mammalian cells in culture is in the form of a noncovalent dimer (unpublished results).
Figure 3.
Analysis of VEGF-DΔNΔC by silver staining, Western blotting, and a bioassay to assess binding to VEGFR-2. (A) Silver stain (Left) and Western blot analysis (Right) of VEGF-DΔNΔC arising from affinity purification (fraction 3). Samples analyzed by silver staining were the fraction containing VEGF-DΔNΔC (+) and a control fraction arising from affinity chromatography of the conditioned medium from cells transfected with expression vector lacking VEGF-D coding sequences (−). Western blot analysis was carried out by using the fraction containing VEGF-DΔNΔC with mAb M2 or a control isotype-matched antibody (Neg). Molecular mass markers (kDa) are indicated. (B) Analysis of VEGF-DΔNΔC using the VEGFR-2 bioassay to assess binding to the extracellular domain of VEGFR-2. Bioassay cells (104 cells) were washed to remove IL-3 and incubated with the recombinant VEGF-DΔNΔC in fraction 3 from the M2 affinity chromatography (VEGF-DΔNΔC). The negative controls were cell culture medium without added growth factor (Medium Alone) and fraction 3 from affinity chromatography of the conditioned medium from cells transfected with expression vector lacking VEGF-D coding sequences (Vector). The positive control was a series of doubling dilutions of mouse VEGF164 from an initial concentration of 100 ng/ml (VEGF164). VEGF-DΔNΔC was also tested against Ba/F3 cells expressing a chimeric receptor consisting of the extracellular domain of Tie2 and the transmembrane and cytoplasmic domains of EpoR (VEGF-DΔNΔC: Tie2/EpoR). All of the fractions used for the assays were tested at an initial concentration of 10% in cell culture medium followed by doubling dilutions. The concentration of VEGF-DΔNΔC at 10% dilution of fraction 3 was 300 ng/ml. Cells were incubated for 48 h, and cell proliferation was then quantitated by the addition of [3H]thymidine and measuring the amount incorporated over a 4-h period. Assays were carried out in duplicate and error bars denote 1 SD.
VEGF-DΔNΔC was tested in a bioassay to assess binding of ligands to the extracellular domain of mouse VEGFR-2. The VEGFR-2 bioassay involved the use of a cell line expressing a chimeric receptor consisting of the extracellular ligand-binding domain of mouse VEGFR-2 and the transmembrane and cytoplasmic domains of the EpoR. The chimeric receptor had been transfected into the Ba/F3 cell line, a line that is IL-3-dependent. When another receptor capable of delivering a growth stimulus is transfected into Ba/F3 cells, the cells can be rescued in the absence of IL-3 by the specific growth factor that activates that receptor. The expression of the VEGFR-2/EpoR chimeric receptor in Ba/F3 cells means that when a ligand binds to the extracellular domain of VEGFR-2, signaling from the cytoplasmic domain of the EpoR will result, causing the cells to survive and proliferate in the absence of IL-3 (27).
VEGF-DΔNΔC (from fraction 3) was able to induce DNA synthesis in the bioassay cell line at a concentration of 50 ng/ml (Fig. 3B). In comparison, the corresponding fraction from control enrichments of conditioned medium from cells that had been transfected with the expression vector pEFBOSSFLAG showed no activity, as did the cell culture medium without addition of affinity-purified material. The dependence of the response to VEGF-DΔNΔC on the extracellular domain of VEGFR-2 was demonstrated by the finding that VEGF-DΔNΔC was unable to promote DNA synthesis in Ba/F3 cells expressing a chimeric receptor (S.A.S. and A.S. Runting, unpublished results) consisting of the extracellular domain of mouse Tie2 (29) and the cytoplasmic domain of EpoR (Fig. 3B). As expected, mouse VEGF164 strongly induced DNA synthesis in the bioassay cells. These results clearly demonstrate that VEGF-DΔNΔC can bind to the extracellular domain of VEGFR-2.
VEGF-D Activates VEGFR-2 and VEGFR-3.
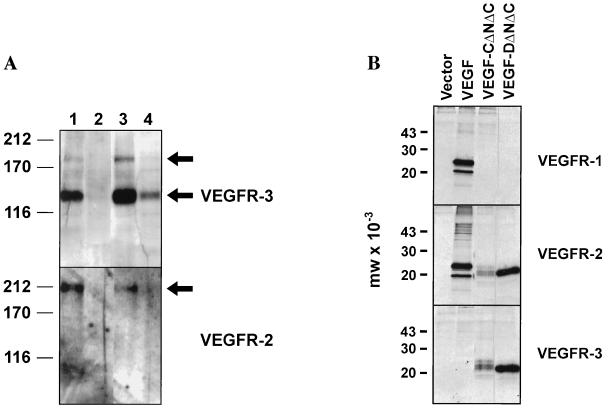
The ability of VEGF-D to induce tyrosine phosphorylation of human VEGFR-3 and human VEGFR-2 (KDR) was examined. For this purpose, VEGF-DΔNΔC and full-length VEGF-D, both with C-terminal histidine tags, were expressed in Baculovirus and designated VEGF-DΔNΔC-H6 and full-length VEGF-D-H6, respectively. The resulting cell supernatants containing these recombinant proteins were used to stimulate either NIH 3T3 cells expressing human VEGFR-3 (30) or porcine aortic endothelial cells expressing human VEGFR-2 (31). After stimulation, cells were lysed and VEGFR-3 or VEGFR-2 were immunoprecipitated and examined by Western blot analysis with phosphotyrosine-specific mAbs. Both VEGF-DΔNΔC-H6 and full-length VEGF-D-H6 stimulated tyrosine phosphorylation of the 125-kDa processed mature form of VEGFR-3, and phosphorylation of the 195-kDa unprocessed form, stimulated by VEGF-DΔNΔC-H6, was also detected (Fig. 4A Upper, lanes 3 and 4). Stronger phosphorylation was reproducibly generated with cell supernatants containing VEGF-DΔNΔC-H6 than with those containing full-length VEGF-D-H6 (results with duplicate supernatants not shown). As expected, the positive control supernatant (VEGF-CΔNΔC) containing a polypeptide consisting of the region of VEGF-C that is related in primary structure to VEGF-DΔNΔC strongly induced tyrosine phosphorylation of VEGFR-3 (Fig. 4A Upper, lane 1).
Figure 4.
Activation of the receptor tyrosine kinases VEGFR-3 and VEGFR-2 by VEGF-D and precipitation of VEGF-D by soluble VEGFR-Ig fusion proteins. (A) Activation of VEGFR-2 and VEGFR-3 by VEGF-D. Recombinant proteins expressed in Baculovirus were used to stimulate NIH 3T3 cells expressing VEGFR-3 or porcine aortic endothelial cells expressing VEGFR-2. After stimulation, cells were lysed and receptors were immunoprecipitated with receptor-specific antibodies and analyzed by Western blotting with phosphotyrosine-specific antibodies. (Upper) Activation of VEGFR-3 with VEGF-CΔNΔC (lane 1), supernatant from uninfected cells (lane 2), VEGF-DΔNΔC-H6 (lane 3), and full-length VEGF-D-H6 (lane 4). Arrows denote the positions of the phosphorylated proteolytically processed 125-kDa form and the unprocessed 195-kDa form of VEGFR-3. (Lower) Activation of VEGFR-2 with VEGF-CΔNΔC (lane 1), supernatant from uninfected cells (lane 2), VEGF-DΔNΔC-H6 (lane 3), and full-length VEGF-D-H6 (lane 4). The arrow denotes the position of the phosphorylated VEGFR-2. The positions of molecular mass markers, in kDa, are shown to the left. (B) Precipitation of VEGF-D by soluble VEGFR-Ig fusion proteins. Precipitation of labeled VEGF165, VEGF-CΔNΔC, and VEGF-DΔNΔC by VEGFR-1-Ig, VEGFR-2-Ig, and VEGFR-3-Ig was carried out. The fusion proteins used for the precipitations, denoted by the name of the VEGFR involved, are shown to the right. The lane marked Vector denotes results of precipitations from medium derived from cells transfected with expression vector lacking sequence encoding VEGFs.
VEGF-DΔNΔC-H6 also stimulated tyrosine phosphorylation of VEGFR-2 (Fig. 4A Lower, lane 3). This finding was confirmed by using supernatants from two independent transfections with the VEGF-DΔNΔC-H6 plasmid construct (data for duplicate supernatant not shown). As expected, positive control supernatant containing human VEGF-CΔNΔC induced phosphorylation of VEGFR-2 (Fig. 4A Lower, lane 1). In contrast to the relatively strong phosphorylation induced by VEGF-DΔNΔC-H6, phosphorylation of VEGFR-2 induced by full-length VEGF-D-H6 was undetectable.
Purified baculoviral VEGF-DΔNΔC-H6 was tested in the bioassay to assess binding to the extracellular domain of VEGFR-2 (assay described above) and was shown to induce levels of stimulation similar to those of VEGF-DΔNΔC produced in COS cells (data not shown).
VEGF-D Binds to Soluble VEGFR-2 and VEGFR-3 Extracellular Domains.
To further assess the interactions between VEGF-D and VEGFRs, VEGF-DΔNΔC was tested for its capacity to bind to soluble Ig fusion proteins containing the extracellular domains of human VEGFR-1, human VEGFR-2, and human VEGFR-3. The fusion proteins, designated VEGFR-1-Ig, VEGFR-2-Ig, and VEGFR-3-Ig, were expressed, coupled to protein A-Sepharose, and incubated with metabolically labeled human VEGF-DΔNΔC, human VEGF-CΔNΔC, and human VEGF165. Labeled proteins that bound to the fusion proteins were analyzed by SDS/PAGE (Fig. 4B). A polypeptide of the size for VEGF-DΔNΔC (∼22 kDa) was precipitated by VEGFR-2-Ig and VEGFR-3-Ig from the medium of cells expressing VEGF-DΔNΔC. In contrast, no protein of this size was precipitated from the same medium by VEGFR-1-Ig. Essentially the same results were observed for precipitation of VEGF-CΔNΔC. As expected, a predominant polypeptide of approximately 24 kDa was precipitated by VEGFR-1-Ig and VEGFR-2-Ig from the medium of cells expressing VEGF165 but was not precipitated by VEGFR-3-Ig. No labeled polypeptides were precipitated by the three fusion proteins from the medium of cells transfected with the expression vector lacking sequences encoding VEGFs. These data indicate that VEGF-DΔNΔC can bind to VEGFR-2 and VEGFR-3 but not to VEGFR-1.
VEGF-D Is Mitogenic for Endothelial Cells.
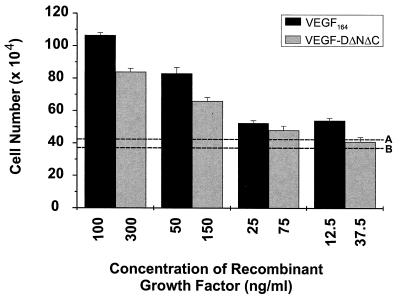
The mitogenic capacity of VEGF-DΔNΔC was tested by using BAEs. BAEs were exposed to VEGF-DΔNΔC for 3 days, and the cells were then dissociated with trypsin and counted. Purified mouse VEGF164 was used as positive control (Fig. 5). Dilution of VEGF-DΔNΔC (fraction 3) into the cell culture medium to give a concentration of 300 ng/ml led to an approximately 2-fold greater increase in the number of BAEs after 3 days in comparison to the same dilution of a fraction that was affinity-purified from the conditioned culture medium of cells transfected with pEFBOSSFLAG. Clearly, VEGF-DΔNΔC is mitogenic for BAEs but is approximately 5-fold less potent than VEGF164 (Fig. 5).
Figure 5.
Mitogenic effect of VEGF-DΔNΔC on BAEs. BAEs were treated with VEGF-DΔNΔC arising from affinity chromatography or a negative control fraction from affinity chromatography of the conditioned medium from cells transfected with expression vector lacking VEGF-D coding sequences. The fraction containing VEGF-DΔNΔC was diluted in cell culture medium containing 5% fetal bovine serum to give the concentrations of growth factor shown. The VEGF-DΔNΔC concentration of 300 ng/ml was achieved by a 1:10 dilution. The positive control was HPLC-purified mouse VEGF164 in cell culture medium at the concentrations shown. Dotted line A indicates the response to a 1:10 dilution of the negative control fraction. Dotted line B indicates the response of the cells to the cell culture medium alone. BAEs were seeded at a density of 104 cells per well for all assays. The results are expressed as the mean ± 1 SD.
DISCUSSION
The complexity of endothelial cell development indicates that its regulation must involve many growth- and tissue-specific differentiation factors that can bind and activate endothelial cell receptors. Therefore, we carried out computer searches of the nucleic acid databases that led to identification of VEGF-D. The fact that VEGF-D is most closely related to VEGF-C is apparent for two reasons. (i) The VEGF homology domain of VEGF-D is much more closely related to that found in VEGF-C than to those of the other family members. (ii) Only VEGF-C has long N- and C-terminal extensions as does VEGF-D. The presence of these extensions in VEGF-C and VEGF-D defines a subfamily of the VEGFs.
The similarity between VEGF-D and VEGF-C exists also at the functional level, because receptor-binding studies reported herein demonstrated that VEGF-D exhibits receptor specificities similar to those of VEGF-C. It has been shown previously that the biosynthesis of VEGF-C involves proteolytic processing, similar to that proposed for platelet-derived growth factor BB (33), which generates a mature polypeptide consisting of the VEGF homology domain (16). The mature VEGF-C polypeptide can bind and activate both VEGFR-2 and VEGFR-3. Our preliminary immunoprecipitation studies of VEGF-D biosynthesis suggested that this protein is processed in a fashion similar to that of VEGF-C. On the basis of this information, we have expressed the region of VEGF-D, corresponding to fully processed VEGF-C, and have shown that this polypeptide (VEGF-DΔNΔC) binds to the extracellular domains of VEGFR-2 and VEGFR-3 and induces tyrosine phosphorylation of these receptors. VEGF-DΔNΔC does not bind to the extracellular domain of VEGFR-1, as is the case for VEGF-CΔNΔC.
Because the receptor-binding specificities of VEGF-D and VEGF-C are similar, an understanding of the biological function of VEGF-C may provide insight into the role played by VEGF-D. It has previously been suggested that VEGF-C may act in a paracrine fashion to regulate angiogenesis of the lymphatic vasculature (14), because VEGFR-3 is strongly expressed by lymphatic endothelium (13). Consistent with this hypothesis was the finding that transgenic mice in which VEGF-C cDNA was expressed in skin exhibited lymphatic endothelial proliferation and vessel enlargement immediately under the skin (34). Given that VEGF-D can also activate VEGFR-3, it is possible that VEGF-D could be involved in the regulation of the growth and/or differentiation of lymphatic endothelium. Because approaches for studying lymphatic endothelium in vitro are extremely limited, it will be important to develop animal models for VEGF-D to study its biological function.
VEGFR-2 (35) and VEGFR-3 (13) are widely expressed during early embryonic development in endothelial cells. Later, in the fetus, VEGFR-3 becomes largely confined to lymphatic endothelial cells (13), whereas VEGFR-2 is expressed in vascular endothelial cells (35). Because the mature forms of both VEGF-C and VEGF-D can activate both of these receptors, VEGF-C and VEGF-D could play roles in coordinating the development of vascular and lymphatic endothelia. It is conceivable that expression of VEGF-C or VEGF-D at a particular site in an embryo could serve to attract the growth of both vascular and lymphatic endothelia, whereas expression of VEGF, a protein that activates VEGFR-2 but not VEGFR-3 (15, 35), would only attract the growth of vascular endothelium. Thus, changes in the levels of expression of the genes for VEGF family members could serve to modulate the abundance of different types of vessels in tissue.
The expression of the gene for mouse VEGF-D is induced by the transcription factor c-fos (18). Continuous expression of c-fos under the control of ubiquitous promoters in transgenic mice induced formation of osteosarcomas (36) and tumors generated from c-fos-deficient cells failed to undergo malignant progression (37). Such experiments have indicated an essential role for c-fos in malignant tumor progression. It will be important to determine whether high levels of VEGF-D gene expression occur in tumors that overexpress c-fos and whether or not VEGF-D can thereby contribute to the malignant tumor phenotype by promoting vascular and/or lymphatic angiogenesis.
Previously, little was known about the molecules that regulate endothelial cell development, although the importance of VEGF in blood vessel formation in the early embryo had been well established (5, 6). The recent discoveries of additional members of the VEGF family, namely, VEGF-B (11), VEGF-C (12, 15), and VEGF-D, have brought to light three more proteins involved in development of the endothelium. The current challenge is to delineate the specific functions of the VEGF family members in blood vessel growth during embryonic development and disease progression in adults.
Acknowledgments
We thank Clare McFarlane for provision of mammalian expression vectors; Katri Pajusola, Yuji Gunji, and Eija Korpelainen for plasmids encoding VEGFR-Ig fusion proteins; Andrew Runting for help with computing; and Helen Cooper and Tony Burgess for critically reading the manuscript. We gratefully acknowledge the support of the National Health and Medical Research Council of Australia.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BAE, bovine aortic endothelial cell; EpoR, erythropoietin receptor; EST, expressed sequence tag; IL-3, interleukin 3; PlGF, placenta growth factor; VEGF, vascular endothelial growth factor; VEGFR, VEGF receptor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ000185).
References
- 1.Risau W. Nature (London) 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J, Shing Y. J Biol Chem. 1992;267:10931–10934. [PubMed] [Google Scholar]
- 3.Risau W. FASEB J. 1995;9:926–933. [PubMed] [Google Scholar]
- 4.Thomas K A. J Biol Chem. 1996;271:603–606. doi: 10.1074/jbc.271.2.603. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P, Ferreira V, Breier G, Pollofeyt S, Keickens L, Gertenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declerq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Nature (London) 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 6.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea K S, Powel-Braxton L, Hillan K J, Moore M W. Nature (London) 1996;380:439–443. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 7.Senger D R, Galli S J, Dvorak A M, Perruzzi C A, Harvey V S, Dvorak H F. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- 8.Maglione D, Guerriero V, Viglietto G, Delli Bovi P, Persico M G. Proc Natl Acad Sci USA. 1991;88:9267–9271. doi: 10.1073/pnas.88.20.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J E, Chen H H, Winer J, Houck K A, Ferrara N. J Biol Chem. 1994;269:25646–25654. [PubMed] [Google Scholar]
- 10.DiSalvo J, Bayne M L, Conn G, Kwok P W, Trivedi P G, Soderman D D, Palisi T M, Sullivan K A, Thomas K A. J Biol Chem. 1995;270:7717–7723. doi: 10.1074/jbc.270.13.7717. [DOI] [PubMed] [Google Scholar]
- 11.Olofsson B, Pajusola K, Kaipainen A, von Euler G, Joukov V, Saksela O, Orpana A, Pettersson R F, Alitalo K, Eriksson U. Proc Natl Acad Sci USA. 1996;93:2576–2581. doi: 10.1073/pnas.93.6.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joukov V, Pajusola K, Kaipainen A, Chilov D, Lahtinen I, Kukk E, Saksela O, Kalkkinen N, Alitalo K. EMBO J. 1996;15:290–298. [PMC free article] [PubMed] [Google Scholar]
- 13.Kaipainen A, Korhonen J, Mustonen T, van Hinsbergh V W, Fang G H, Dumont D, Breitman M, Alitalo K. Proc Natl Acad Sci USA. 1995;92:3566–3570. doi: 10.1073/pnas.92.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kukk E, Lymboussaki A, Taira S, Kaipainen A, Jeltsch M, Joukov V, Alitalo K. Development (Cambridge, UK) 1996;122:3829–3837. doi: 10.1242/dev.122.12.3829. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Gray A, Yuan J, Luoh S M, Avraham H, Wood W I. Proc Natl Acad Sci USA. 1996;93:1988–1992. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joukov V, Sorsa T, Kumar V, Jeltsch M, Claesson-Welsh L, Cao Y, Saksela O, Kalkkinen N, Alitalo K. EMBO J. 1997;16:3898–3911. doi: 10.1093/emboj/16.13.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald N Q, Hendrickson W A. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 18.Orlandini M, Marconcini L, Ferruzzi R, Oliviero S. Proc Natl Acad Sci USA. 1996;93:11675–11680. doi: 10.1073/pnas.93.21.11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson W R. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 20.Lennon G G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 21.Weindel K, Marme D, Weich H A. Biochem Biophys Res Commun. 1992;183:1167–1174. doi: 10.1016/s0006-291x(05)80313-4. [DOI] [PubMed] [Google Scholar]
- 22.Maglione D, Guerriero V, Viglietto G, Ferraro M G, Aprelikova O, Alitalo K, Del Vecchio S, Lei K J, Chou J Y, Persico M G. Oncogene. 1993;8:925–931. [PubMed] [Google Scholar]
- 23.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dignam S S, Case S T. Gene. 1990;88:133–140. doi: 10.1016/0378-1119(90)90024-l. [DOI] [PubMed] [Google Scholar]
- 25.Fung M C, Hapel A J, Ymer S, Cohen D R, Johnson R M, Campbell H D, Young I G. Nature (London) 1984;307:233–237. doi: 10.1038/307233a0. [DOI] [PubMed] [Google Scholar]
- 26.Aruffo A, Seed B. Proc Natl Acad Sci USA. 1987;84:8573–8577. doi: 10.1073/pnas.84.23.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacifici R E, Thomason A R. J Biol Chem. 1994;269:1571–1574. [PubMed] [Google Scholar]
- 28.D’Andrea A D, Lodish H F, Wong G G. Cell. 1989;57:277–285. doi: 10.1016/0092-8674(89)90965-3. [DOI] [PubMed] [Google Scholar]
- 29.Sato T N, Qin Y, Kozak C A, Audus K L. Proc Natl Acad Sci USA. 1993;90:9355–9358. doi: 10.1073/pnas.90.20.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pajusola K, Aprelikova O, Armstrong E, Morris S, Alitalo K. Oncogene. 1993;8:2931–2937. [PubMed] [Google Scholar]
- 31.Waltenberger J, Claesson Welsh L, Siegbahn A, Shibuya M, Heldin C H. J Biol Chem. 1994;269:26988–26995. [PubMed] [Google Scholar]
- 32.Breier G, Albrecht U, Sterrer S, Risau W. Development (Cambridge, UK) 1992;114:521–532. doi: 10.1242/dev.114.2.521. [DOI] [PubMed] [Google Scholar]
- 33.Ostman A, Thyberg J, Westermark B, Heldin C H. J Cell Biol. 1992;118:509–519. doi: 10.1083/jcb.118.3.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeltsch M, Kaipainen A, Joukov V, Meng X, Lakso M, Rauvala H, Swartz M, Fukumura D, Jain R K, Alitalo K. Science. 1997;276:1423–1425. doi: 10.1126/science.276.5317.1423. [DOI] [PubMed] [Google Scholar]
- 35.Millauer B, Wizigmann Voos S, Schnurch H, Martinez R, Moller N P, Risau W, Ullrich A. Cell. 1993;72:835–846. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- 36.Grigoriadis A E, Wang Z Q, Wagner E F. Trends Genet. 1995;11:436–441. doi: 10.1016/s0168-9525(00)89142-8. [DOI] [PubMed] [Google Scholar]
- 37.Saez E, Rutberg S E, Mueller E, Oppenheim H, Smoluk J, Yuspa S H, Spiegelman B M. Cell. 1995;82:721–732. doi: 10.1016/0092-8674(95)90469-7. [DOI] [PubMed] [Google Scholar]