Abstract
The caspases are cysteine proteases that have been implicated in the execution of programmed cell death in organisms ranging from nematodes to humans. Many members of the Bcl-2 family, including Bcl-xL, are potent inhibitors of programmed cell death and inhibit activation of caspases in cells. Here, we report a direct interaction between caspases and Bcl-xL. The loop domain of Bcl-xL is cleaved by caspases in vitro and in cells induced to undergo apoptotic death after Sindbis virus infection or interleukin 3 withdrawal. Mutation of the caspase cleavage site in Bcl-xL in conjunction with a mutation in the BH1 homology domain impairs the death-inhibitory activity of Bcl-xL, suggesting that interaction of Bcl-xL with caspases may be an important mechanism of inhibiting cell death. However, once Bcl-xL is cleaved, the C-terminal fragment of Bcl-xL potently induces apoptosis. Taken together, these findings indicate that the recognition/cleavage site of Bcl-xL may facilitate protection against cell death by acting at the level of caspase activation and that cleavage of Bcl-xL during the execution phase of cell death converts Bcl-xL from a protective to a lethal protein.
Many cell death stimuli, including growth factor withdrawal, receptor ligation, drug treatment, and virus infection, have been shown to activate the cellular family of cysteine proteases known as caspases (1–3). This and other accumulating evidence strongly indicates that the caspases are crucial for the execution phase of apoptosis. Caspases are synthesized as zymogens that require proteolytic cleavage to generate active enzyme subunits. These activating cleavage events can be carried out by other caspases and are thought to represent a major regulatory step in the apoptosis pathway. In addition to caspases themselves, several other target substrates of the caspases have been identified, including transcription factors, kinases, enzymes involved in DNA repair, cytoskeletal proteins, and others (1, 2, 4–6). However, the role of these cleavage events in cell death remains to be fully elucidated.
By unknown mechanisms, the activation of caspases is inhibited by Bcl-2 and Bcl-xL (7–14). Bcl-2 family members may facilitate inhibition of caspase activation through one or more of their assigned functions, such as pore-formation, homo- and heterodimerization, and other protein–protein interactions (12, 15–17). A potential mechanism by which Bcl-xL may regulate caspases is suggested by the recent identification of several proteins, p28Bap31 (18), MRIT (19), and Caenorhabditis elegans CED-4 (9, 14, 20–23), that physically bridge Bcl-xL with caspases. However, a direct interaction between Bcl-xL and caspases has not been reported previously.
Although the overall amino acid identity among Bcl-2 family members is low, there are several conserved motifs, BH1 to BH4 (24). The BH1 and BH2 domains of Bcl-2 and Bcl-xL are important for heterodimerization with other Bcl-2 family members (25, 26). Although the BH1 and BH2 domains are required for anti-death activity, heterodimerization apparently is not required to inhibit apoptosis (26). The death-inducing members of the Bcl-2 family, Bak and Bax, contain a BH3 domain that appears to be sufficient to induce cell death (27, 28). Of interest, this domain also is conserved in the anti-death family members Bcl-2 and Bcl-xL. A fourth conserved domain located near the N terminus, BH4, forms an α-helix and is required for the anti-apoptotic activity of Bcl-2 (29, 30).
Here, we demonstrate that Bcl-xL is a caspase substrate. Caspase cleavage of Bcl-xL in the loop region releases a C-terminal product that lacks the BH4 homology domain and potently induces cell death. Mutation of the caspase cleavage site inhibits release of the proapoptotic fragment, correlating with improved cell viability. However, the cleavage site mutation causes a loss of anti-death activity when combined with a mutation in the BH1 domain that previously was shown to block heterodimerization with Bax and Bak without significantly affecting anti-death activity (26). Therefore, the loss of anti-death activity by this double mutant suggests that, before proteolysis of Bcl-xL, the caspase cleavage site is involved in inhibition of cell death, perhaps by inhibiting caspase activation.
MATERIALS AND METHODS
Plasmid Constructions.
Rat bcl-x (GenBank accession no. U10579) was amplified from rat dorsal root ganglia cDNA by PCR by using primers for human 5′ and 3′ bcl-x untranslated sequences (31). cDNAs encoding human interleukin (IL) 1, cowpox virus crmA, human caspase-3, baculovirus P35, murine Bax, human Bcl-2 and Bcl-xL, and rat Bcl-xL and Bcl-xS were expressed from the pSG5 vector (Stratagene). Mutations in human Bcl-xL were generated by recombinant PCR and confirmed by DNA sequencing. The coding sequences for Bcl-xL amino acids 62–233 plus an initiation codon were amplified by PCR to construct mutant ΔN61. The Bcl-xL Δloop mutant was obtained from Craig Thompson (University of Chicago) (32) and lacks amino acids 26–83. D61A/BH1(mt1) and Δloop/BH1(mt1) were constructed by ligating DNA fragments from the D61A or Δloop mutants and BH1(mt1) (26).
Cleavage Assays.
Cleavage reactions contained 3 μl of 35S-labeled in vitro translation mixture and 1 μl (30 units) of purified caspase-1 [provided by Susan Molineaux, Merck; 1 unit generates 1 pmol 7-amino-4-methylcoumarin (AMC)/min by using saturating substrate (Ac-YVAD-AMC, Peptides International) at 25°C] or 2 μl of purified caspase-3 (provided by Kristine Kikly, SmithKline; 1 μl produces 2.5 × 103 relative fluorescence units/min using DEVD-AMC). DTT was added to a final concentration of 10 mM, and caspase reaction buffer (100 mM Hepes pH 7.5/10% sucrose/0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) was added to bring the total volume of each reaction to 20 μl. After incubation at 30°C for 6 h, the labeled proteins were analyzed by SDS/PAGE and fluorography.
Cleavage Site Mapping.
Glutathione S-transferase–Bcl-xL fusion protein lacking the transmembrane domain was purified from Escherichia coli, and 5 μg was incubated overnight with caspase-1 as described above. The cleavage product was separated by SDS/PAGE and transferred to Immobilon Psq poly(vinylidene difluoride) membrane (Millipore) for microsequencing.
Cell Extracts.
S/M and IP extracts were prepared as described (33). Each reaction contained 10 mM DTT (final concentration), 0.5 μl of in vitro translation mixture, and 7.5 units of caspase-1 or 3 μl S/M or IP extract. Caspase reaction buffer was used to bring each reaction to a total volume of 5 μl. The reactions were preincubated with or without inhibitors for 15 minutes at 37°C before addition of in vitro translation mixture and further incubated at 37°C for 6 h before analysis by SDS/PAGE and fluorography.
Immunoblotting.
BHK cells were transfected by using lipofectamine (Life Technologies, Grand Island, NY), with total DNA concentrations being held constant by addition of control plasmid pSG5. At 24 h after transient transfection, cells were lysed in RIPA buffer (150 mM NaCl/1.0% Nonidet P-40/0.5% deoxycholate/0.1% SDS/50 mM Tris, pH 8.0) and 50 μg of protein (quantitated by bicinchoninic acid assay; Pierce) were separated by SDS/PAGE. Proteins were transferred to nitrocellulose and probed with anti-Bcl-x mAb 2A1 (34) (provided by Craig Thompson) and detected by using enhanced chemiluminescence (Amersham). COS-1 cells were harvested in RIPA buffer and Ba/F3 cells were harvested in 2X Laemmli sample buffer (35) and 50 μg (COS-1) or 20 μg (Ba/F3) of protein per sample were separated by 17.5% Tricine-SDS/PAGE (36), transferred to Immobilon P membrane (Millipore), and immunoblotted with 2A1 antibody and enhanced chemiluminescence.
Cell Death Assays.
Recombinant Sindbis virus vectors were generated as described (26). Cells were infected with 10 plaque-forming units of virus per cell, and cell viability was determined at 48 h postinfection by trypan blue exclusion.
Ba/F3 stable cell lines were established as described (35, 37). Cells were electroporated with pBabePuro (38) alone or with pSG5–Bcl-xL constructs and selected in 2 μg/ml puromycin. Individual clones were analyzed by immunoblotting, and lines with similar levels of protein expression were selected for further analysis. At various times after IL-3 withdrawal, cell viability was determined by propidium iodide staining and FACScan flow cytometry.
The lacZ transfection viability assays were performed as described (39). BHK cells were transfected with 0.5 μg each of lacZ reporter plasmid pCH110 and the Bcl-x plasmids indicated by using lipofectamine with equal amounts of total DNA per sample. At 24 h after transfection, cells were fixed and stained with 5-bromo-4-chloro-3-indolylβ-d-galactoside. Cell viability was measured by counting blue cells in 10 high power fields and scoring normal vs. apoptotic morphology.
RESULTS
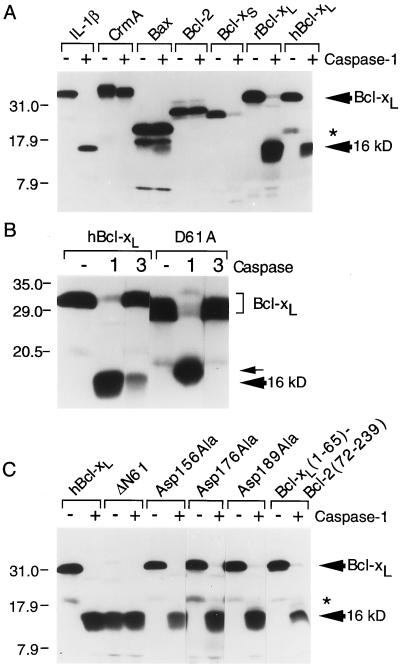
To determine whether Bcl-xL could be digested by caspases, in vitro-translated proteins were treated with purified active enzymes. When radioactively labeled Bcl-2, Bax, Bcl-xS, and Bcl-xL proteins were treated with caspase-1 (IL-β converting enzyme), only Bcl-xL (both rat and human) and Bcl-xS were proteolyzed (Fig. 1A). A stable digestion product of Bcl-xS was not detected, but cleavage of Bcl-xL by caspase-1 produced a stable 16-kDa fragment. As expected, caspase-1 also cleaved its natural substrate proIL-1β and the cowpox virus caspase inhibitor CrmA (which is cleaved only 38 amino acids from the C terminus) (40). Cleavage of Bcl-xL was inhibited by the caspase-1 inhibitors YVAD-CMK and iodoacetate but not by leupeptin, phenylmethylsulfonyl fluoride, or pepstatin, which is consistent with the inhibitor profile of caspase-1 (data not shown). Bcl-xL also was susceptible to cleavage by caspase-3 (Fig. 1B).
Figure 1.
In vitro cleavage of Bcl-xL by caspases. (A-C) The indicated in vitro-translated 35S-labeled proteins were incubated with the indicated recombinant caspases and analyzed by SDS/PAGE and fluorography. The D61A mutation causes an altered migration in SDS/PAGE (see Discussion). Met45 of human Bcl-xL (an Arg in rat Bcl-xL) is presumed to give rise to an internal initiation product marked by an asterisk throughout.
The caspase-1 cleavage site in Bcl-xL was determined by N-terminal sequencing of the 16-kDa cleavage product derived from digestion of the glutathione S-transferase–Bcl-xL fusion protein with caspase-1 (data not shown). The resulting peptide sequence placed the cleavage site after an aspartate in the sequence His-Leu-Ala-Asp61 of Bcl-xL. An aspartate residue at the P1 position is consistent with other known caspase substrates as well as caspase inhibitors (2, 41, 42). To verify the cleavage site mapping, the aspartate at position 61 was mutated to alanine. This D61A mutation rendered Bcl-xL resistant to digestion by caspase-3 in vitro (Fig. 1B). The D61A mutation also abolished cleavage after residue 61 by caspase-1, but an alternate cleavage occurred at a more N-terminal site producing a slightly larger product (Fig. 1B, small arrow). Deletion of the Bcl-xL loop domain (amino acids 26–83) completely abolished cleavage of Bcl-xL by caspase-1 and -3 (data not shown). In contrast to Asp61, mutation of the three most C-terminal aspartate residues of Bcl-xL (Asp 156, 176, 189) had no effect on caspase-1 cleavage, indicating that the 16-kDa fragment was derived from an N-terminal cleavage event rather than both N- and C-terminal processing (Fig. 1C). Further confirmation of the Asp61 cleavage site was obtained by digestion of a Bcl-xL/Bcl-2 chimera containing the N-terminal 65 amino acids (including the cleavage site) of Bcl-xL and the C-terminal 167 amino acids of Bcl-2, which also produced a single 16-kDa fragment (Fig. 1C). In addition, the translation product of a Bcl-xL clone lacking the first 61 amino acids (ΔN61) comigrated with the caspase-1 cleavage product of wt Bcl-xL and was resistant to caspase-1 digestion (Fig. 1C).
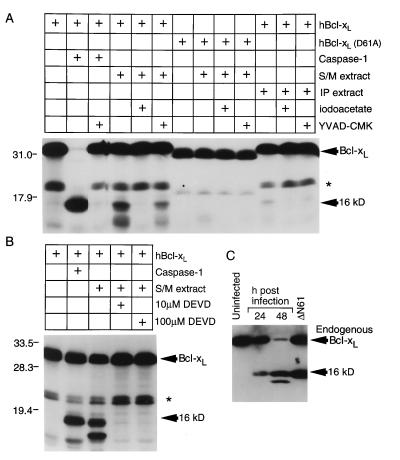
Cytoplasmic extracts from cells undergoing the early stages of apoptosis have been useful in recapitulating many of the important events occurring during the execution phase of cell death. Addition of apoptotic S/M extracts to radiolabeled Bcl-xL produced a 16-kDa product and a smaller unidentified fragment, but a control cell extract (IP) prepared from nonapoptotic cells had little effect (Fig. 2A). Processing of Bcl-xL by the S/M extract was inhibited by 10 μM DEVD-CHO (peptide aldehyde) (Fig. 2B) and 2 mM iodoacetate but only partially inhibited by 100 μM YVAD-CMK (peptide chloromethylketone) (Fig. 2A). These results are consistent with a previous report that S/M extracts do not contain any detectable caspase-1 activity but instead contain abundant caspase-3 activity (43). However, these experiments do not distinguish between cleavage by caspase-3 and cleavage by other DEVD-CHO-sensitive enzymes. The S/M extract failed to cleave the D61A mutant (Fig. 2A), indicating that the S/M protease cleaves Bcl-xL at the same site recognized by purified caspases in vitro. The alternate, more N-terminal site recognized by recombinant caspase-1 was not cleaved by the S/M extract (compare with Fig. 1B).
Figure 2.
Cleavage of Bcl-xL by apoptotic cell extracts and in cells induced to undergo apoptotic cell death. (A) An in vitro translation mix containing 35S-labeled wt or mutant Bcl-xL protein was incubated with recombinant caspase-1, apoptotic cell extract (S/M), or control extract (IP) with or without inhibitors and analyzed by SDS/PAGE and fluorography. Induction of apoptotic morphology in HeLa cell nuclei by the S/M extract and lack of apoptosis with the IP extract was confirmed by 4′,6-diamidino-2-phenylindole staining (data not shown). (B) An independent experiment in which cleavage of in vitro-translated Bcl-xL by the S/M extract was inhibited by Ac-DEVD-CHO. ∗, See legend to Fig. 1. (C) Cleavage of endogenous Bcl-xL in COS-1 cells after Sindbis virus infection. Virus-infected or ΔN61-transfected cell lysates were immunoblotted with 2A1 antibody.
To determine whether endogenous Bcl-xL was cleaved after a physiological death stimulus, COS-1 cells were infected with Sindbis virus (44, 45) and immunoblotted for Bcl-x. A prominent 16-kDa polypeptide that comigrated with transfected ΔN61 was detected only in virus-infected cell lysates (Fig. 2C). A smaller fragment also was detected at late times similar to that observed with cell extracts (compare with Fig. 2 A and B). Increased cleavage of endogenous Bcl-xL correlated with reduced cell viability at 24 (75.7%) and 48 h (19.9%) postinfection compared with mock-infected cells (95.5%). Thus, cleavage of endogenous Bcl-xL by endogenous caspases was observed after induction of apoptosis by virus infection.
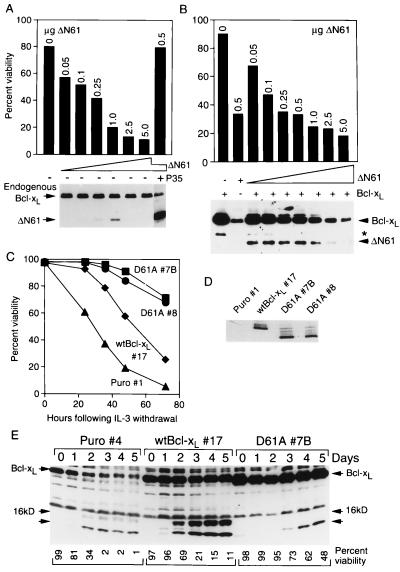
To determine what effect caspase cleavage has on Bcl-xL function, a construct lacking the first 61 amino acids of Bcl-xL (ΔN61) was transfected into BHK cells along with a lacZ reporter plasmid. Transfection of ΔN61 potently induced dose-dependent apoptotic cell death characterized by cell shrinkage and membrane blebbing (Fig. 3A and data not shown). Transfection of only 50 ng of ΔN61 plasmid reduced the viability of transfected cells from 80 to 57% despite the fact that the ΔN61 protein was undetectable by immunoblot analysis. In fact, the ΔN61 protein could only be detected in a narrow window of transfected DNA concentrations, presumably because of its potent pro-death activity. The difficulty in detecting ΔN61 Bcl-xL in some cells may explain why caspase cleavage of Bcl-xL previously went undetected. Of interest, cell death induced by ΔN61 was blocked completely by cotransfection with baculovirus P35. Although P35 may also block cleavage of Bcl-xL (not assayed in this experiment), P35 blocks cell death downstream of ΔN61 (Fig. 3A), suggesting that ΔN61 may accelerate cell death by amplifying the caspase cascade. Furthermore, co-transfection of P35 resulted in significantly increased accumulation of the ΔN61 protein, supporting the hypothesis that the potent pro-death activity of ΔN61 leads to elimination of those cells that express it, making the cleavage product difficult to detect. (The overall levels of endogenous Bcl-xL remained largely unchanged because the majority of cells were not transfected.)
Figure 3.
The C-terminal cleavage product of Bcl-xL induces apoptosis, and mutation of the Bcl-xL cleavage site enhances anti-death function. (A) BHK cells were transfected with a lacZ reporter plasmid plus increasing amounts of the ΔN61 mutant with or without P35 (0.5 μg), and viability of transfected (blue) cells was determined 24 h later. The results are representative of two independent experiments. Shown below is an immunoblot of similarly transfected cells. (B) BHK cells were cotransfected with a lacZ reporter plasmid, 0.5 μg of wt Bcl-xL and increasing amounts of the ΔN61 mutant or with ΔN61 alone (0.5 μg) and viabilities were determined as in A. The corresponding immunoblot is shown below. (C) Ba/F3 cell lines expressing the indicated proteins were induced to undergo apoptosis by withdrawal of IL-3 as detected by propidium iodide staining. The results shown are the mean ± SD (hidden by the symbols) for a representative experiment performed in triplicate. (D) Equal amounts of protein from the indicated Ba/F3 cell lines were analyzed by immunoblotting; this light exposure does not detect endogenous Bcl-xL. The D61A mutation causes an altered migration on gels (see Discussion). (E) The indicated Ba/F3 cell lines were immunoblotted with 2A1 antibody at the indicated days after IL-3 withdrawal. Cell viabilities determined by propidium iodide staining for this experiment are indicated below.
In contrast to P35, cotransfected wild-type (wt) Bcl-xL (0.5 μg) had little effect on killing induced by ΔN61 (compare viabilities in Fig. 3B with 3A). The increased accumulation of ΔN61 protein at low DNA concentrations probably reflects the increased levels of full length substrate available for digestion. However, the failure of increased levels of cleavage product to further reduce cell viability indicates that the ratio of full length to cleaved Bcl-xL may be important in determining susceptibility to cell death.
To provide evidence that the Bcl-xL cleavage fragment produced by endogenous caspases accelerates cell death, IL-3-dependent murine hematopoietic (Ba/F3) cells were stably transfected with wt Bcl-xL or a mutant of Bcl-xL in which the cleavage site Asp was mutated to Ala. Clones expressing similar amounts of Bcl-xL protein (Fig. 3D) were tested for their ability to block death induced by IL-3 withdrawal. Although wt Bcl-xL protected cells compared with vector alone (Puro), two cell lines expressing the cleavage site mutant D61A exhibited significantly enhanced cell survival compared with wt Bcl-xL (Fig. 3C). Immunoblot analysis of control and transfected Ba/F3 cells at increasing times after IL-3 withdrawal demonstrated that both endogenous and transfected Bcl-xL were cleaved to a 16-kDa fragment and a smaller fragment that may correspond to the smaller fragment observed with S/M extracts (Fig. 3E). The appearance of the cleavage product also correlated with reduced levels of full length protein (more evident in lighter exposures in the case of Bcl-xL #17). The 16-kDa fragment was derived by cleavage at Asp61 because mutation of Asp61 in the D61A #7B cell line abolished production of the 16-kDa fragment although a smaller fragment was produced at later time points (Fig. 3E).
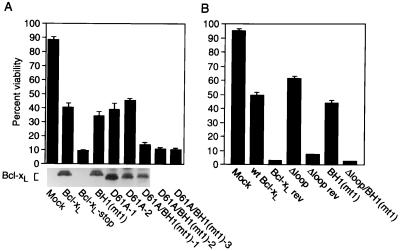
The presence of a caspase cleavage site in Bcl-xL, a protein that inhibits caspase activation, also raises the possibility that, before cleavage, Bcl-xL could potentially function as a pseudosubstrate to directly inhibit caspases. The presence of Bcl-xL in the same protein complex with caspases fuels this hypothesis (9, 18, 19). However, the purified glutathione S-transferase–Bcl-xL protein did not inhibit cleavage of recombinant proIL-1β by purified caspase-1 (data not shown). Furthermore, the finding that the loop domain is dispensable for the anti-death activity of Bcl-xL (32) indicates that the cleavage site is not required to inhibit cell death at least in some scenarios. Therefore, to explore the possibility that Asp61 has a role in anti-death activity, the cleavage site mutation D61A was studied in the context of a mutant BH1 domain. Bcl-xL mutant 1 (mt1) has two point mutations in BH1 that abolish heterodimerization with Bax and Bak but do not significantly alter anti-death activity (26). Although neither the cleavage site mutation nor the BH1 (mt1) mutation impaired anti-death activity, the combination of these mutations in the same protein abolished anti-death activity when assayed with the Sindbis virus vector system (Fig. 4A). Results obtained with three independent D61A/BH1(mt1) constructs were similar to a negative control in which a premature stop codon was inserted near the N terminus of Bcl-xL (Bcl-xL-stop). The wt and mutant Bcl-xL proteins were expressed at similar levels in infected cells as assessed by immunoblotting (Fig. 4A and data not shown). Thus, Asp61 is vital for protection in this assay when BH1 is mutated. If this is the case, then deletion of the loop domain, which normally enhances anti-death activity, also would be expected to abolish anti-death activity when combined with the BH1 mutation (mt1). As predicted, the Δloop/BH1(mt1) mutant failed to protect BHK cells from virus-induced apoptosis whereas the Δloop mutation alone provided consistently enhanced protection (62%) compared with wt Bcl-xL (50%) (Fig. 4B) (32).
Figure 4.
The caspase cleavage site in Bcl-xL is important for anti-death activity in the context of a BH1 mutation. (A and B) BHK cells were mock-infected or infected with recombinant Sindbis viruses expressing wt Bcl-xL or the indicated Bcl-xL mutants. The results are the mean ± SE of three to four independent experiments, counting at least 500 cells per sample. (A) An immunoblot of similarly infected cell lysates. The antibody does not detect the loop deletion mutant. Control recombinant viruses expressing Bcl-xL in the reverse orientation are indicated as “rev.”
DISCUSSION
The evidence presented here indicates that endogenous and overexpressed Bcl-xL is cleaved by endogenous caspases during cell death induced by IL-3 withdrawal or virus infection. Cleavage of Bcl-xL in the loop domain effectively removes the N-terminal BH4 homology domain, converting Bcl-xL into a potent pro-death molecule. Like Bcl-xL, Bcl-2 also is cleaved by caspases in the loop domain to release a potently pro-apoptotic C-terminal cleavage product (46). Activation of Bcl-xL and Bcl-2 pro-death activity after cleavage could potentially serve as a feed-forward mechanism for executing the cell. It has been reported that deletion of the Bcl-2 BH4 domain (amino acids 4–29) destroyed its anti-death function (29, 30). Surprisingly, this mutant failed to exhibit pro-death activity (28), suggesting the possibility that pro-death activity of the cleavage product may be suppressed in some situations.
The sequence of the Bcl-2 cleavage site (DAGD) more closely resembles the site recognized by class II caspases whereas the Bcl-xL cleavage site does not resemble other known sites at the P4 position. Further evidence that Bcl-2 and Bcl-xL are cleaved by distinct proteases comes from the observation that, although Bcl-xL is cleaved by caspase-1 in vitro, Bcl-2 is not susceptible to digestion by caspase-1 (see Fig. 1A). Furthermore, Fas-induced cell death in Jurkat cells is associated with significant cleavage of endogenous Bcl-2, but endogenous Bcl-xL is cleaved much less efficiently in the same cells (ref. 46 and data not shown). Thus, the release of pro-apoptotic cleavage products of Bcl-2 and Bcl-xL is likely to be regulated by the particular subset of activated caspases.
A caspase cleavage site also is conserved in CED-9, the Bcl-2 homolog found in C. elegans, except that cleavage of CED-9 by the C. elegans CED-3 protease occurs between the BH4 domain and an N-terminal extension not found in mammalian Bcl-2 or Bcl-xL (47). Therefore, the cleavage product of CED-9 retains the BH4 domain and accordingly lacks pro-death activity. The N-terminal 65-aa extension on CED-9 may represent the functional equivalent of the loop domain found in Bcl-xL and Bcl-2 but lacking from CED-9. Thus, the conservation of a caspase cleavage site in mammalian and C. elegans Bcl-2 family proteins does not appear to be for the purpose of releasing a proapoptotic fragment. Perhaps the commonality lies with the anti-death activity of the caspase cleavage site. Mutation of the CED-9 cleavage site in combination with a mutation in the BH1 domain of CED-9 abolishes anti-death activity of CED-9 (47) similar to that demonstrated here for Bcl-xL. The requirement for both the cleavage site and an intact BH1 domain could imply that Bcl-xL has at least two functional domains, one in BH1, perhaps required for ion channel activity and/or protein–protein interactions, and another in the loop domain involving caspase interaction. An alternative possibility is that the loop domain and the BH1 domain participate in the same function or that the loop domain has an alternate activity involving proteins other than caspases. These potential mechanisms are not mutually exclusive.
Genetic evidence from C. elegans suggests that normal surviving cells contain CED-3 protease activity that is held in check by CED-9 (48). Likewise, it is possible that the loop region of Bcl-xL functions to inhibit basal levels of caspase activity, perhaps through a direct interaction with the caspase active site that is facilitated by a bridging protein such as MRIT, BAP31, a mammalian CED-4 homolog (49), or by other mechanisms. Mutation of the cleavage site Asp61 in Bcl-xL caused an altered migration of Bcl-xL in SDS/PAGE, whereas mutation of several other Asp residues had no effect (compare Fig. 1C to Figs. 1B, 2A, 3D, and 4A). Similarly, mutation of the reactive site Asp in P35 and CrmA also causes altered SDS/PAGE migration (data not shown), perhaps due to alterations in protein structure.
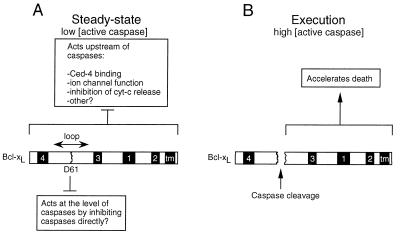
In summary, our data suggest that the caspase cleavage site in Bcl-xL serves as a double-edged sword. Before receiving a death stimulus, it functions in conjunction with BH1 to inhibit basal caspase activity (Fig. 5A). When the cell receives a death stimulus and more caspases become activated, caspase cleavage at Asp61 converts Bcl-xL into a pro-death molecule (Fig. 5B) that further amplifies the caspase cascade (see Fig. 3A). Thus, there may be a delicate balance between anti-death and pro-death activities contained within the same protein, which is regulated by the levels of active caspases in the cell.
Figure 5.
A model for regulation of cell death by Bcl-xL. (A) Under normal conditions (low concentrations of active caspases), Bcl-xL may have two separate anti-death functions, one that acts through factors binding to the BH1 domain and acts upstream of caspases (7–13, 26) and the other, localized in the loop domain, that functions to directly inhibit low levels of active caspases. (B) When the death stimulus has overwhelmed the anti-death activity of Bcl-xL, higher levels of active caspases accumulate, and cleavage of the loop region of Bcl-xL leads to the accumulation of Bcl-xL cleavage product that may accelerate the death process. Numbered boxes refer to the BH domains in Bcl-xL, and tm indicates the transmembrane domain.
Acknowledgments
We thank Suchada Kwunyeun and Beverly Plunkett for technical assistance and Brian Chang, Christine Canman, Kristine Kikly, Yuri Lazebnik, Susan Molineaux, Craig Thompson, and Junying Yuan for reagents. R.J.C. is an American Cancer Society Fellow and D.G.K. is supported by the National Institutes of Health Medical Scientist Training Program. This research was supported by grants from the National Institutes of Health and the Muscular Dystrophy Association (J.M.H.).
ABBREVIATIONS
- IL
interleukin
- wt
wild-type
References
- 1.Fraser A, Evan G. Cell. 1996;85:781–784. doi: 10.1016/s0092-8674(00)81005-3. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi A, Earnshaw W C. Curr Opin Genet Dev. 1996;6:50–55. doi: 10.1016/s0959-437x(96)90010-6. [DOI] [PubMed] [Google Scholar]
- 3.Nava V E, Rosen A, Veliuona M A, Clem R J, Levine B, Hardwick J M. J Virol. 1998;72:452–459. doi: 10.1128/jvi.72.1.452-459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Zou H, Slaughter C, Wang X. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 5.Tan X Q, Martin S J, Green D R, Wang J Y J. J Biol Chem. 1997;272:9613–9616. doi: 10.1074/jbc.272.15.9613. [DOI] [PubMed] [Google Scholar]
- 6.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry T J, Kirschner M W, Koths K, Kwiatkowski D J, Williams L T. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 7.Zamzami N, Susin S A, Marchetti P, Hirsch T, Gomez-Monterrey I, Castedo M, Kroemer G. J Exp Med. 1996;183:1533–1544. doi: 10.1084/jem.183.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 9.Chinnaiyan A M, O’Rourke K, Lane B R, Dixit V M. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 11.Chinnaiyan A M, Orth K, O’Rourke K, Duan H, Poirier G G, Dixit V M. J Biol Chem. 1996;271:4573–4577. doi: 10.1074/jbc.271.9.4573. [DOI] [PubMed] [Google Scholar]
- 12.Minn A J, Velez P, Schendel S L, Liang H, Muchmore S W, Fesik S W, Fill M, Thompson C B. Nature (London) 1997;385:353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 13.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 14.Seshagiri S, Miller L K. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 15.Schendel S L, Xie Z, Montal M O, Matsuyama S, Montal M, Reed J C. Proc Natl Acad Sci USA. 1997;94:5113–5118. doi: 10.1073/pnas.94.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod J-J, Mazzei G, Maundrell K, Gambale F, Sadoul R, Martinou J-C. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 17.Reed J C. Nature (London) 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 18.Ng F W H, Nguyen M, Kwan T, Branton P E, Nicholson D W, Cromlish J A, Shore G C. J Cell Biol. 1997;139:327–338. doi: 10.1083/jcb.139.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han D K M, Chaudhary P M, Wright M E, Friedman C, Trask B J, Riedel R T, Baskin D G, Schwartz S M, Hood L. Proc Natl Acad Sci USA. 1997;94:11333–11338. doi: 10.1073/pnas.94.21.11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irmler M, Hofmann K, Vaux D, Tschopp J. FEBS Lett. 1997;406:189–190. doi: 10.1016/s0014-5793(97)00271-8. [DOI] [PubMed] [Google Scholar]
- 21.Wu D, Wallen H D, Nunez G. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 22.Spector M S, Desnoyers S, Hoeppner D J, Hengartner M O. Nature (London) 1997;385:653–655. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 23.James C, Gschmeissner S, Fraser A, Evan G I. Curr Biol. 1997;7:246–252. doi: 10.1016/s0960-9822(06)00120-5. [DOI] [PubMed] [Google Scholar]
- 24.Kroemer G. Nat Med. 1997;3:614–620. doi: 10.1038/nm0697-614. [DOI] [PubMed] [Google Scholar]
- 25.Yin X-M, Oltvai Z N, Korsmeyer S J. Nature (London) 1994;369:321–323. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
- 26.Cheng E H-Y, Levine B, Boise L H, Thompson C B, Hardwick J M. Nature (London) 1996;379:554–556. doi: 10.1038/379554a0. [DOI] [PubMed] [Google Scholar]
- 27.Chittenden T, Flemington C, Houghton A B, Ebb R G, Gallo G J, Elangovan B, Chinnadurai G, Lutz R J. EMBO J. 1995;14:5589–5596. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter J J, Parslow T G. J Biol Chem. 1996;271:8521–8524. doi: 10.1074/jbc.271.15.8521. [DOI] [PubMed] [Google Scholar]
- 29.Borner C, Martinou I, Mattmann C, Irmler M, Schaerer E, Martinou J-C, Tschopp J. J Cell Biol. 1994;126:1059–1068. doi: 10.1083/jcb.126.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter J J, Bond B L, Parslow T G. Mol Cell Biol. 1996;16:877–883. doi: 10.1128/mcb.16.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boise L H, Gonz‡lez-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nuñez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 32.Chang B S, Minn A J, Muchmore S W, Fesik S W, Thompson C B. EMBO J. 1997;16:968–977. doi: 10.1093/emboj/16.5.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazebnik Y A, Cole S, Cooke C A, Nelson W G, Earnshaw W C. J Cell Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boise L H, Minn A J, Noel P J, June C H, Thompson C B. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 35.Canman C E, Gilmer T M, Coutts S B, Kastan M B. Genes Dev. 1995;9:600–611. doi: 10.1101/gad.9.5.600. [DOI] [PubMed] [Google Scholar]
- 36.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 37.Palacios R, Steinmetz M. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 38.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 40.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 41.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:1–20. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 42.Xue D, Horvitz R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 43.Lazebnik Y A, Kaufmann S H, Desnoyers S, Poirier G G, Earnshaw W C. Nature (London) 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 44.Levine B, Huang Q, Isaacs J T, Reed J C, Griffin D E, Hardwick J M. Nature (London) 1993;361:739–742. doi: 10.1038/361739a0. [DOI] [PubMed] [Google Scholar]
- 45.Lewis J, Wesselingh S L, Griffin D E, Hardwick J M. J Virol. 1996;70:1828–1835. doi: 10.1128/jvi.70.3.1828-1835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng E H-Y, Kirsch D G, Clem R J, Ravi R, Kastan M B, Bedi A, Ueno K, Hardwick J M. Science. 1997;278:1966–1968. doi: 10.1126/science.278.5345.1966. [DOI] [PubMed] [Google Scholar]
- 47.Xue D, Horvitz H R. Nature (London) 1997;390:305–308. doi: 10.1038/36889. [DOI] [PubMed] [Google Scholar]
- 48.Shaham S, Horvitz H R. Genes Dev. 1996;10:578–591. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- 49.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]