Abstract
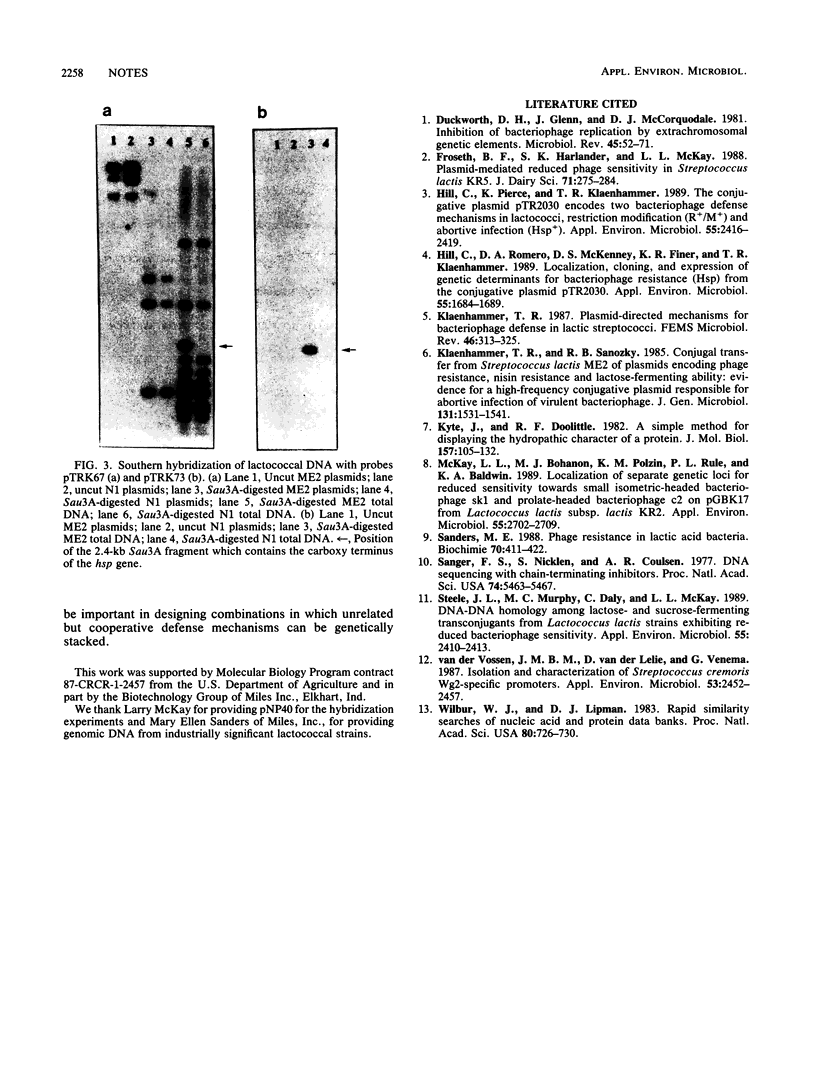
The lactococcal plasmid pTR2030 encodes resistance to bacteriophage attack via two mechanisms, an abortive-infection mechanism, designated Hsp, and a restriction and modification system. We present the complete sequence of the hsp structural gene. The gene is 1,887 base pairs in length and encodes a protein with a predicted molecular mass of 73.8 kilodaltons. The upstream region was cloned in a promoter-screening vector and shown to direct the constitutive expression of the cat-86 gene. An internal probe was used to determine the distribution of the hsp sequence in industrially significant lactococcal strains and to evaluate its relatedness to another lactococcal plasmid implicated in an abortive-infection-type mechanism, pNP40. No homology was detected, suggesting that this gene is not widely distributed in lactococci. Therefore, there are at least two independent abortive-infection genotypes in lactococci.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Duckworth D. H., Glenn J., McCorquodale D. J. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol Rev. 1981 Mar;45(1):52–71. doi: 10.1128/mr.45.1.52-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froseth B. R., Harlander S. K., McKay L. L. Plasmid-mediated reduced phage sensitivity in Streptococcus lactis KR5. J Dairy Sci. 1988 Feb;71(2):275–284. doi: 10.3168/jds.S0022-0302(88)79555-7. [DOI] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Bohanon M. J., Polzin K. M., Rule P. L., Baldwin K. A. Localization of Separate Genetic Loci for Reduced Sensitivity towards Small Isometric-Headed Bacteriophage sk1 and Prolate-Headed Bacteriophage c2 on pGBK17 from Lactococcus lactis subsp. lactis KR2. Appl Environ Microbiol. 1989 Oct;55(10):2702–2709. doi: 10.1128/aem.55.10.2702-2709.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E. Phage resistance in lactic acid bacteria. Biochimie. 1988 Mar;70(3):411–422. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J. L., Murphy M. C., Daly C., McKay L. L. DNA-DNA homology among lactose- and sucrose-fermenting transconjugants from Lactococcus lactis strains exhibiting reduced bacteriophage sensitivity. Appl Environ Microbiol. 1989 Sep;55(9):2410–2413. doi: 10.1128/aem.55.9.2410-2413.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]