Abstract
Tumor necrosis factor (TNF) can induce a variety of cellular responses at low picomolar concentrations. This is in apparent conflict with the published dissociation constants for TNF binding to TNF receptors in the order of 100–500 pM. To elucidate the mechanisms underlying the outstanding cellular sensitivity to TNF, we determined the binding characteristics of TNF to both human TNF receptors at 37°C. Calculation of the dissociation constant (Kd) from the association and dissociation rate constants determined at 37°C revealed a remarkable high affinity for TNF binding to the 60-kDa TNF type 1 receptor (TNF-R1; Kd = 1.9 × 10−11 M) and a significantly lower affinity for the 80-kDa TNF type 2 receptor (TNF-R2; Kd = 4.2 × 10−10 M). The high affinity determined for TNF-R1 is mainly caused by the marked stability of ligand–receptor complexes in contrast to the transient interaction of soluble TNF with TNF-R2. These data can readily explain the predominant role of TNF-R1 in induction of cellular responses by soluble TNF and suggest the stability of the TNF–TNF receptor complexes as a rationale for their differential signaling capability. In accordance with this reasoning, the lower signaling capability of homotrimeric lymphotoxin, compared with TNF, correlates with a lower stability of the lymphotoxin–TNF-R1 complex at 37°C.
Tumor necrosis factor (TNF) is a pleiotropic cytokine that is a major mediator of immunological and pathophysiological reactions. As with many other cytokines, the concentration of TNF found in body fluids in pathophysiological situations is usually very low (1, 2). Various cellular responses of cultured cells can be initiated by low picomolar or even femtomolar concentrations of TNF (3–6). Because the binding of TNF to its cell surface receptors is a prerequisite for TNF responses, high-affinity binding sites for TNF with affinity constants in the same range should be assumed.
Two distinct membrane receptors for TNF (TNF-Rs) with apparent molecular weights of 55–60 kDa (TNF-R1) and 70–80 kDa (TNF-R2) have been identified and molecularly cloned (for review, see ref. 7). Both TNF-Rs have been objects of intense physiological and biochemical investigations. Likewise, the ligand binding properties of the TNF-Rs have been extensively studied (8–11) although a detailed analysis became feasible only after the molecular cloning of the two individual receptor molecules. Equilibrium binding studies with 125I-labeled TNF (125I-TNF) at 0°C defined high-affinity binding of TNF to both TNF-Rs with Kd values of approximately 300–600 pM for TNF-R1 and 70–200 pM for TNF-R2 (12–18).
Although most cell lines and primary tissues coexpress both receptor types, cellular responses to the soluble 17-kDa form of TNF seems to be dominated by the interaction with TNF-R1 (19–21). On the other hand, we have shown recently in different cellular systems that TNF-R2 can be strongly stimulated by the 26-kDa transmembrane form of TNF (transmembrane TNF) rather than by the soluble form, suggesting that transmembrane TNF is the prime physiological activator of TNF-R2 (22). Accordingly, the contribution of TNF-R2 to cellular responses induced by soluble TNF appears to be mainly of supportive or modulating nature. In a recent report the rapid kinetics of TNF–TNF-R2 association and dissociation have been taken as a basis to postulate a model termed “ligand passing” in which the ligand bound to TNF-R2 may be passed over to TNF-R1 to enhance TNF-R1 signaling (23). As a consequence, the effective association rate of TNF to TNF-R1 in cells coexpressing both receptors would be significantly increased, explaining the low TNF concentrations sufficient to trigger cellular responses. However, comparable prominent cellular sensitivities for TNF could be also demonstrated with cells that only express TNF-R1 (24) leaving open the question of how such low TNF concentrations might be effective.
In the present work, we examined the binding affinities of TNF-R1 and TNF-R2 for soluble TNF at 37°C, thus resembling physiological conditions. Determination of the dissociation constant (Kd) by means of measurement of the respective association/dissociation rates at 37°C revealed a significantly higher affinity of TNF binding to TNF-R1 when compared with binding studies at 0°C. From these data it is evident that TNF-R1, and not TNF-R2, is the high-affinity receptor for soluble TNF, giving a rationale for the predominant role of TNF-R1 in cellular responses induced by soluble TNF.
MATERIALS AND METHODS
Cells and Reagents.
Recombinant human TNF (2 × 107 units/mg) and lymphotoxin (LT; 6 × 107 units/mg) were provided by Knoll (Ludwigshafen, Germany) and Bender MedSystems (Vienna, Austria). The biological activities of TNF and LT were routinely controlled by using L929 cells as described (25). The molar concentrations used herein refer to trimeric TNF and LT, respectively. The human cell lines HeLa and U937 were obtained from the American Type Culture Collection and the murine MethA sarcoma cell line (26) was kindly provided by M. Clauss (Max-Planck-Institute, Bad Nauheim, Germany). Cells were grown in RPMI 1640 medium supplemented with 5% heat-inactivated fetal calf serum and 10 mM glutamine (all from Biochrom, Berlin). The human rhabdomyosarcoma cell line KYM-1 was supplied by M. Sekiguchi (University of Tokyo, Japan) and maintained in Click’s RPMI medium (Biochrom) containing 10% fetal calf serum. The generation and specificity of Fab fragments of the TNF-R1-specific mAb H398 (19) and polyclonal rabbit anti-human TNF-R2-specific IgG (17) have been described.
Binding Experiments.
TNF and LT were labeled with Na125I (Amersham) by the chloramine-T method to specific activities of 0.5–1.5 × 108 cpm/μg with retention of 30–60% of biologic activity for TNF and 20–50% for LT, as determined by the KYM-1 cell cytotoxicity assay (17). For association kinetic experiments, the radiolabeled cytokines were prewarmed (37°C) in 75 μl of Hanks’ balanced salt solution (HBSS) containing 2% fetal calf serum and 20 mM sodium azide (binding buffer) on a mixture of dibutyl phthalate and bis(2-ethylhexyl) phthalate oil (Sigma), optimized for each different cell line. The reaction was started by the addition of 75 μl of the respective prewarmed (37°C) cell suspension and stopped by centrifugation of the tubes (Sarstedt) for 20 sec in a microcentrifuge (14,000 × g; Eppendorf). The time needed for the separation of the cells from the binding medium containing the free radiolabeled ligand had been determined to be 6 sec and was included in the total reaction time. After centrifugation the samples were immediately placed on dry ice and cell-bound radioactivity was determined by cutting the tips of the tubes and measuring the radioactivity in a counter (Berthold, Wildbad, Germany). Nonspecific binding was determined in parallel by measuring the binding of the radiolabeled cytokine in the presence of a 200-fold molar excess of unlabeled TNF.
For dissociation kinetic experiments, cells were preincubated in the presence or absence of competing receptor-specific Fab fragments (30 μg/ml) for 1 hr at 4°C followed by incubation with the 125I-labeled cytokine for 1 hr at 4°C in the absence or presence of an excess of unlabeled TNF for the determination of the nonspecific binding in a total volume of 60 μl of binding buffer. Subsequently, 50 μl of the cell suspension was suspended in 100 μl of prewarmed binding buffer (37°C) containing a 200-fold excess of unlabeled TNF on a layer of phthalate oil for several time periods. The time point at which the cells were resuspended was taken as the start of the reaction. Cell-bound radioactivity was determined by centrifugation as described above.
Kinetic Data Analysis.
Net association and dissociation rates were calculated from the data after subtraction of nonspecific binding by nonlinear regression by using the program graphpad prism. The respective dissociation constants (Kd) were calculated from the ratio of the dissociation rate (koff) and the association rate constants (kon) with kon = (kobs − koff)/[L], where kobs is the net association rate and [L] is the concentration of the respective radiolabeled ligand. If reversible binding is assumed, the receptor occupancy was calculated for a given ligand concentration according to the Langmuir adsorption isotherm with occupancy = [L]/([L] + Kd).
Cytotoxicity Assay.
Cells were seeded in 96-well microtiter plates (Greiner, Nürtingen, Germany) at 3 × 104 cells per well in the presence of cycloheximide (1 μg/ml) and the various reagents were added. After 18 hr, culture supernatants were discarded, and the cells were washed once with phosphate-buffered saline followed by crystal violet staining (20% methanol/0.5% crystal violet) for 15 min. The wells were washed with water and air-dried. The dye was eluted with methanol for 15 min and optical density at 550 nm was measured with a R5000 ELISA plate reader (Dynatech).
RESULTS
Determination of the Kinetic TNF Binding Parameters at 37°C.
The binding affinities of TNF for the two TNF-Rs have been extensively studied by steady-state binding experiments at 0°C. To clarify the interaction of TNF with the two TNF-Rs under more physiological conditions, we performed binding studies at 37°C. However, because equilibrium saturation isotherms at 37°C are affected by secondary events such as ligand-mediated receptor internalization and receptor turnover, the ligand–receptor affinity (expressed as dissociation constant, Kd) was calculated from the independently determined association and dissociation rates. To study the kinetics of TNF binding to each of the two receptors independently, we used HeLa cells, almost exclusively expressing TNF-R1 (19, 20), and KYM-1 cells, which predominantly (>90%) express TNF-R2 (17).
The off rates of 125I-TNF from TNF-R1 and TNF-R2 were determined by displacement experiments. After HeLa and KYM-1 cells had been incubated with radiolabeled ligand for 1.5 hr at 0°C, the cells were transferred quickly to prewarmed (37°C) culture medium containing an excess of unlabeled ligand, and then the decrease of cell bound 125I-TNF was followed. These experiments were performed within a period of less than 8 min in which ligand internalization was controlled to account for less than 15% of total ligand binding. All values are corrected for nonspecific binding. The data in Fig. 1A show that TNF rapidly dissociates from TNF-R2 with a half-life time of 1.1 min (koff value 0.631 ± 0.241 min−1, n = 4), a result consistent with a previous study (22). In contrast, the dissociation of TNF from TNF-R1 (Fig. 1A) was found to be extremely slow (t½ = 33.2 min; koff value = 0.021 ± 0.009 min−1, n = 7).
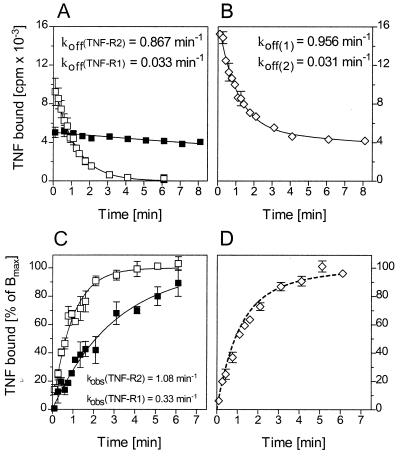
Figure 1.
Dissociation and association kinetics of 125I-TNF binding at 37°C. (A) Approximately 4 × 105 KYM-1 cells (□, approximately 30,000 TNF-R2 per cell) and HeLa cells (▪, approximately 3,000 TNF-R1 per cell) were preincubated with anti-TNF-R1 antibody H398 (KYM-1) or left untreated (HeLa) and then incubated with 300 pM of 125I-TNF for 1 hr at 4°C. Subsequently, dissociation of 125I-TNF from TNF-Rs at 37°C was followed by dilution of the cells in a 200-fold excess of unlabeled TNF and determination of cell-bound radioactivity by centrifugation of the cells through a layer of phthalate oil at the indicated time points. Shown is a typical experiment (KYM-1, n = 4; HeLa, n = 7) with 31,535 cpm and 2,017 cpm of specific binding at time 0 for KYM-1 and HeLa cells, respectively. Nonspecific binding was 3,482 cpm and 182 cpm, respectively, and has been subtracted. (B) For association kinetics 4.5 × 105 HeLa cells (▪) and KYM-1 cells (□) were incubated at 37°C for different times with 300 pM 125I-TNF. Unbound ligand was then removed by centrifugation of the cells through phthalate oil and cell bound radioactivity was measured. A typical experiment (KYM-1, n = 4; HeLa, n = 7) is shown with 41,640 cpm and 2,451 cpm of maximum specific binding for KYM-1 and HeLa cells, respectively. Nonspecific binding determined in the presence of 60 nM unlabeled TNF has been subtracted. The continuous curves passing through the data in A and B were calculated from the best-fit parameter values by using single-exponential time-dependency curves.
Fig. 1B shows the association kinetics of 125I-TNF and demonstrates that the kinetics of TNF binding to both TNF-Rs is very rapid with a somewhat slower association to TNF-R1 than to TNF-R2. Net association rates (kobs) of 0.34 min−1 and 1.49 min−1 for TNF-R1 and TNF-R2, respectively, were calculated from the data by best-fit single-exponential time-dependency plots. Similar association kinetic experiments were performed with TNF concentrations of 0.1–1.6 nM (data not shown). The resulting experimentally observed net association rates together with the concentration of the iodinated ligand [L] and the respective dissociation rate constants were used to calculate the association rate constants. The derived values were 1.1 ± 0.2 × 109 (n = 7) for TNF-R1 and 1.5 ± 0.6 × 109 M−1⋅min−1 (n = 4) for TNF-R2. The association and dissociation rate constants were used to calculate the Kd values, describing the affinities of TNF-R1 and TNF-R2 for TNF. The derived Kd values are 4.2 × 10−10 M for TNF-R2 and 1.9 × 10−11 M for TNF-R1. Most remarkably, at physiological temperature the affinity of TNF-R1 for TNF is about 20 times higher when compared with equilibrium binding studies performed at 0°C (see above).
To further corroborate the high affinity of TNF for TNF-R1, we performed two additional experimental approaches to determine the kinetic constants. First, we measured the net association rates of TNF to HeLa cells at 37°C as a function of different TNF concentrations (Fig. 2A Inset). The resulting kobs values were linear dependent of the TNF concentrations used, as expected for a bimolecular reaction (Fig. 2A). Within this plot, the intercept on the ordinate represents the dissociation rate constant and the slope is equal to the association rate constant (27). The resulting mean values from three independent experiments were 0.039 ± 0.012 min−1 for the dissociation rate constant and 1.01 ± 0.10 × 109 M−1·min−1 for the association rate constant and are thus in good agreement with the values obtained in the experiments described above (Fig. 1).
Figure 2.
Alternative methods to determine association and dissociation rate constants. (A) Association experiments with HeLa cells were performed as in Fig. 1 by using various 125I-TNF concentrations (100, 200, 400, 800, and 1,600 pM) (Inset). The obtained net association rate values were plotted against the 125I-TNF concentration initially present in the assay to determine the dissociation rate constant (ordinate intercept) and the association rate constant (slope). Shown is a representative experiment (n = 3). (B) HeLa cells were incubated at 37°C with 250 pM 125I-TNF to obtain the association kinetics shown (▪). After 2, 5, and 7 min of incubation (arrows), aliquots were supplemented with a 200-fold excess of unlabeled TNF in prewarmed binding buffer, and cell-bound radioactivity was followed for an additional 7 min (○). The dotted curves passing through the data represent the calculated dissociation kinetics based on the dissociation rate constants obtained from the experiments shown in Fig. 1.
Second, we followed the dissociation process of TNF after a short-term incubation of cells with the iodinated ligand at 37°C. These experiments were performed to exclude more stringently that the observed slow dissociation kinetics of TNF from TNF-R1 were caused by artefacts as a consequence of the preincubation of the cells with the radiolabeled ligand on ice or the subsequent temperature shift to 37°C. Accordingly, dissociation of 125I-TNF was induced by the addition of an excess of unlabeled TNF after 2, 5, and 7 min of an association time course (Fig. 2B). The data in this experimental set-up confirm the slow dissociation of TNF from TNF-R1.
Thus, these data show that TNF-R1, and not TNF-R2, is the high-affinity receptor for soluble TNF. In addition, these findings emphasize the importance to analyze the cytokine binding parameters under physiological conditions for a determination of the relevant affinity values. What are the reasons for the different Kd values obtained at 0°C and 37°C? First, we have confirmed that both techniques, direct determination by equilibrium binding studies and calculation from kinetic experiments, reveal similar Kd values at 0°C. In particular, the kinetic parameters at 0°C were 0.037 min−1 (koff) and 5.9 × 108 M−1⋅min−1 (kon) for TNF-R2 and 0.0015 min−1 (koff) and 1.3 × 107 M−1⋅min−1 (kon) for TNF-R1. The resulting calculated Kd values were 6 × 10−11 M for TNF-R2 and 1 × 10−10 M for TNF-R1 as compared with 7–20 × 10−11 M for TNF-R2 and 3–6 × 10−10 M for TNF-R1 determined by equilibrium saturation binding studies at 0°C (12–18). Interestingly, the dissociation rate of TNF from TNF-R1 is also very slow at 0°C, which as a consequence requires incubation times of about 40 h to reach equilibrium binding at 0°C. Accordingly, most published Kd values obtained from equilibrium binding studies performed on ice might represent underestimates. In conclusion, the main reason for the different affinity values of TNF-R1 for TNF measured at 0°C and at 37°C lies in the different TNF association rates to TNF-R1 (84-fold). More important, the difference between TNF binding to TNF-R1 and TNF-R2 at 37°C is mainly caused by the distinct dissociation kinetics.
TNF Binding Parameters on TNF-R1/TNF-R2-Coexpressing Cells.
Because most cells coexpress both TNF-Rs, we were interested to know whether some type of receptor interference can be detected that affects the ligand-binding properties described above. Cells of the histiocytic line U937 have been widely studied for TNF–TNF-R interaction, coexpress TNF-R2 and TNF-R1 at a ratio of approximately 2:1 and have been demonstrated to be exclusively sensitive for TNF-mediated effects via TNF-R1 but not TNF-R2 (19, 28, 29). First, we analyzed the TNF binding kinetics of TNF-R1 and TNF-R2 separately by preincubating U937 cells with receptor-specific Fab fragments followed by the measurement of the dissociation and association rates of 125I-TNF at 37°C. The obtained dissociation rates (Fig. 3A) and association rates (Fig. 3C) for TNF-R1 and TNF-R2 on U937 cells were found to be similar to those determined on HeLa and KYM-1 cells, respectively, indicating that the binding properties of TNF-R1 and TNF-R2 are not cell-specific.
Figure 3.
TNF binding kinetics on U937 cells, coexpressing TNF-R1 and TNF-R2. U937 cells (1 × 106 cells) were preincubated with receptor-specific antibodies for 1 hr on ice (A and C) or left untreated (B and D). 125I-TNF dissociation and association kinetics of cells with TNF-R1 (▪), TNF-R2 (□), or both receptors (◊) present were performed at 37°C as described in Fig. 1. The continuous curves passing through the data were calculated from best-fit parameter values by using single-exponential time-dependency curves (A and C) or two-phase-exponential time-dependency curves (B) and the resulting rate constants are given in the graph. The dotted curve passing through the data in D represents the calculated two-phase-exponential association kinetics based on the individual net association rates of TNF-R1 and TNF-R2, respectively, determined in parallel (C). Shown is a typical experiment (n = 3).
Next we analyzed the TNF association and dissociation time course on U937 cells without having blocked either of the TNF-Rs. The dissociation profile was found to be biphasic as best-fit nonlinear regression of the data points yielded optimal results for a two-phase exponential decay with the indicated values for koff(1) and koff(2) (Fig. 3B). Accordingly, two independent binding sites could be observed that most probably represent TNF-R1 and TNF-R2, as concluded from two findings. (i) Both derived koff values correspond closely to the dissociation rate constants when the receptors were analyzed individually (Fig. 3A). (ii) The calculated spans from the nonlinear regression fitting of the two overlapping dissociation profiles [11,370 cpm for span(1) and 5,284 cpm for span(2)] correspond to the amount of 125I-TNF binding to the two individual receptor populations. This could be shown by a competition binding assay performed in parallel at 0°C with the receptor-specific antibodies that competed for 10,510 cpm (TNF-R2-specific antibody) and 5,052 cpm (TNF-R1-specific antibody) of specific 125I-TNF binding. Thus, these kinetic data with U937 cells argue for independent dissociation processes for the two TNF-Rs at 37°C as indicated by studies performed at 0°C (23).
The data of association binding experiments of TNF to U937 cells at 37°C, with both receptors available, did not show a clear two-phases exponential ligand association. This is due to the fact that the individual TNF net association rates of TNF-R1 and TNF-R2 at 37°C are similar (Figs. 1B and 3C). However, a calculated two-phase association curve using the respective net association rates of TNF-R1 and TNF-R2 taken from the data shown in Fig. 3C fits closely to the experimentally measured data points on U937 cells (Fig. 3D), thus, arguing for independent association kinetics of TNF to the individual TNF-Rs at 37°C.
The Bioactivity of LT Correlates with the Dissociation Rate from TNF-R1.
LT, formerly also known as TNF-β, has been shown to bind to both TNF-Rs but exerts a significantly lower biological potency in many TNF-R1-dependent cellular responses of human cells (for example, see refs. 24, 30–32). One example is shown in Fig. 4A, demonstrating an approximately 10-fold lower bioactivity of LT in the induction of cytotoxicity in HeLa cells when compared with TNF. These findings have been so far inconsistent with the similar dissociation constants, derived from equilibrium binding studies at 0°C, for TNF and LT binding to human TNF-R1 (18, 33, 34). With regard to the present investigations, however, it was tempting to speculate about different binding characteristics of LT to TNF-R1 at physiological temperature. In fact, dissociation binding kinetics with the human HeLa cells revealed an about 8-fold faster dissociation of LT from human TNF-R1 (koff(LT) = 0.157 ± 0.029 min−1; n = 6) in comparison to TNF (Figs. 1A and 4C). The difference between LT and TNF in the association rates to human TNF-R1 was less pronounced with net association rates of 0.22 ± 0.01 min−1 (n = 3) for LT and 0.39 ± 0.02 min−1 (n = 3) for TNF when 350 pM of the respective radiolabeled ligand was used. These data suggest that the difference in bioactivities of TNF and LT for human cells resides primarily in the different off rates from TNF-R1.
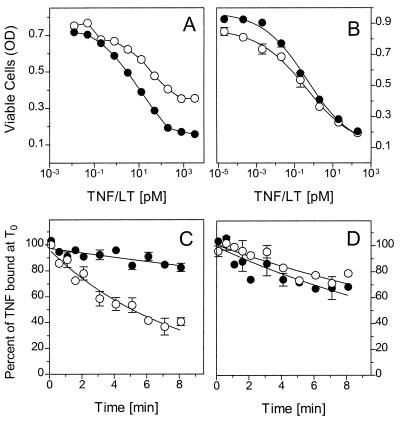
Figure 4.
Comparison of TNF and LTα bioactivities and dissociation rates on human and mouse cells. TNF (•) and LTα (○) were titrated on human HeLa cells (A) and mouse MethA cells (B) in a standard cytotoxicity assay. Note that the full cytotoxic effect of TNF cannot be reached with 2 nM LTα. The dissociation rates of 125I-TNF (•) and 125I-LT (○) from HeLa cells (C) and MethA cells (D) were determined as described in Fig. 1. Shown are typical experiments (HeLa, n = 6; MethA, n = 3).
A prediction from the above reasoning would be that in cellular systems where TNF and LT reveal similar bioactivities both cytokines should display similar kinetic binding parameters at 37°C. Accordingly, we analyzed murine cells for which it has been demonstrated that human LT and human TNF bind only to the murine TNF-R1 and not to the murine TNF-R2 and, more important, reveal comparable bioactivities (3, 31, 34). In fact, in the murine cell lines MethA (Fig. 4) and L929 (data not shown), human LT and human TNF were equally potent in the induction of cytotoxicity (Fig. 4B) and revealed similar dissociation kinetics when analyzed at 37°C (Fig. 4D).
DISCUSSION
In general, cytokines act in a concentration-dependent manner. A prerequisite for intracellular signal initiation is ligand binding to receptors that also follows a similar concentration dependency, suggestive of a causal relationship. Equilibrium binding studies have often been used to characterize the affinity of a given ligand/receptor system. To prevent secondary reactions such as internalization or shedding of the ligand–receptor complexes, these experiments are usually performed at 0°C. However, this experimental approach reveals physiologically relevant data only if association and dissociation kinetics (and thus the Kd value) at 0°C are similar to that at physiological temperature.
For the TNF system, the dissociation constants obtained by equilibrium binding studies at 0°C are typically in the range of 300–600 pM for TNF-R1 and 70–200 pM for TNF-R2 (see above). However, these experimental values are at variance with the extraordinary sensitivity of cells for different TNF responses. For example, the cytotoxic effects of TNF or the activation of the transcription factor NF-κB can be induced by low picomolar or even femtomolar concentrations of TNF (3, 4, 6, 24). In other words, if the Kd values derived from equilibrium binding studies at 0°C are taken as the basis for a calculation of the percentage of TNF-R occupied at such low TNF concentrations, this would mean that significant cellular responses to TNF can already be induced at receptor occupancies of less than 1% (3, 24, 35).
Herein we examined the ligand–receptor binding characteristics of both TNF-Rs at physiological temperature. Different experimental approaches resulted in a consistent picture regarding the physiological ligand affinities of the two TNF-R molecules. In the experiments shown in Fig. 1, we directly determined the association and dissociation kinetics of radiolabeled TNF for each of the receptors independently and calculated the constants kon and koff thereof. The precise mathematical determination of the dissociation rate constant for TNF-R1 by curve fitting was difficult because of the slow kinetics. Therefore, we determined the association and dissociation rates by a second approach, shown in Fig. 2A, measuring the dependency of the association rate from the TNF concentration. Both approaches resulted in comparable values. Thus, these results provide strong evidence that at physiological temperature TNF-R1 and not TNF-R2 is the high-affinity receptor for soluble TNF.
Thus, our study readily explains why most cellular responses to soluble TNF are dominated by TNF-R1, even when considerable numbers of TNF-R2 are coexpressed. When the different affinities of the two TNF-Rs are taken into account, it is feasible that low picomolar concentrations of TNF can efficiently trigger TNF-R1 but the active participation of TNF-R2 as a signaling molecule needs notably higher ligand concentrations. For example, at a given TNF concentration of 10 pM, a fraction of 34% of TNF-R1 would be ligated in equilibrium but only 2% of TNF-R2. It is conceivable that under these conditions TNF-R1 can induce a significant cellular response but TNF-R2 is not able to do so simply because of the limited subcritical ligand interaction.
Recently, studies of the association and dissociation kinetics of TNF at 0°C were taken as the basis to develop a model in which TNF-R2, due to its fast on and off rates, delivers the TNF molecule to TNF-R1 (23, 36). This model might explain why, at low TNF concentrations, TNF-R2-specific antibodies can act as antagonists in certain TNF responses in which only TNF-R1 is functional (23). However, other possible explanations for that phenomenon might also exist such as, e.g., the formation of heteromeric TNF-R complexes (37). On the other hand, the finding that TNF-R1 and TNF-R2 do not affect each other substantially regarding ligand association and dissociation at 37°C (Fig. 3) would rather argue against a dominant proportion of such complexes. In any case, the extraordinary sensitivity of TNF-R1-dependent cellular responses to TNF does not necessarily rely on the presence of “ligand passing” by the TNF-R2 molecule because such sensitivities are also found with cell lines that express TNF-R1 only.
An important aspect in the binding properties of the two TNFR molecules is the remarkable difference between the dissociation rates of TNF from TNF-R1 and TNF-R2 (Fig. 1A). This parameter, representing a true physical constant of the respective ligand–receptor system under reversible conditions, is 30-fold higher for TNF-R2 than for TNF-R1 (Figs. 1 and 2). Accordingly, TNF–TNF-R1 complexes have an extraordinary stability (t½ = 33.2 min), whereas the ligand rapidly dissociates from TNF-R2 (t½ = 1.1 min). These values correspond to mean survival times of individual ligand–receptor complexes of 47.9 min (TNF-R1) and 1.6 min (TNF-R2).
However, under physiological conditions with vital cells, no truly reversible ligand binding occurs due to secondary reactions, mainly internalization and/or shedding of ligand–receptor complexes. Accordingly, the dissociation rates determined in this study might be biased by secondary reactions. In fact, it has been shown that TNF-R1 becomes primarily internalized after ligation, whereas TNF-R2 is rapidly shed (38, 39). However, a principal difference of both receptors in ligand dissociation kinetics was suggested recently by using purified soluble receptor IgG fusion proteins under conditions where reversible binding is given (40). Therefore, irrespective of whether the dissociation constant values obtained in this study with intact cells represent the true physical constants of these ligand/receptor systems at the cellular membrane or whether these values are in part influenced by secondary reactions, it is apparent that they represent the biologically relevant parameters. Accordingly, it is most likely that the vast majority of TNF–TNF-R1 complexes becomes internalized (t½ = 10–20 min; refs. 8 and 41 and unpublished data) rather than dissociates (t½ = 33 min), allowing the efficient formation of signaling complexes that may even persist for prolonged times in an active state intracellularly. On the other hand, TNF-R2 complexes are formed more transiently, possibly limited in signaling because of ligand dissociation and receptor shedding.
Consequently, this reasoning suggests the different stability of ligand–receptor complexes as an important parameter determining TNF-R signaling capacity. This argument is supported by different findings. First, we have recently observed that stabilization of TNF–TNF-R2 complexes by means of a mAb led to a strong enhancement of the signaling potency of soluble TNF via TNF-R2. In parallel, we could show that TNF-R2 can be strongly stimulated by transmembrane TNF rather than by soluble TNF. Both agents (i.e., transmembrane TNF and soluble TNF plus stabilizing antibody) were capable of inducing TNF responses in a quantity and quality that could not be induced by saturating concentrations of soluble TNF (22).
Second, we have herein investigated the kinetic association and dissociation parameters of LT binding to TNF-R1 and the respective mouse homologue (muTNF-R1). For a long time, the physiological role of the soluble homotrimeric LT molecule has been a subject of intensive research and is still a matter of debate (32). This is mainly based on the fact that LT, although more stable under physiological conditions than TNF (34), induces more or less the same cellular response pattern that TNF does but with a considerably lower bioactivity. Again, saturating LT concentrations are unable to induce a response maximum readily obtained with high concentrations of TNF (refs. 32 and 33 and Fig. 4A). On mouse cells, however, human LT exerts a similar bioactivity than human TNF (Fig. 4B). This bioactivity pattern of LT does not correspond to the ligand–receptor affinities determined at 0°C (data not shown) but correlates well with the stability of the respective LT–receptor complexes (Fig. 4 B and C).
How could the observed mean survival time of ligand–receptor complexes determine signaling in a quantitative or even qualitative manner? This might depend on secondary reactions, possibly on the formation of higher receptor aggregates or conformational changes (42). In fact, x-ray crystallographic analyses have suggested that TNF-Rs engage their ligand at a 3:1 stoichiometric ratio (43). In addition, two potential quaternary structural arrangements for the unligated TNF-R1 have been proposed (44). Each of these possibilities would anticipate the interaction of TNF-R1 with the TNF trimer to be a time consuming and temperature-dependent process. Thus, it is reasonable that conformational but also physiologically active processes may influence the ligand binding properties of TNF-R1 giving also a rationale for the different results of TNF binding studies performed at 0°C and 37°C.
Finally, the stability of the ligand–receptor complex will have direct influence on the association of signaling molecules to the cytoplasmic tail of ligated receptors as shown for another member of the TNF family, the APO-1/Fas antigen (45). Because both TNF-Rs are devoid of any enzymatic activity, the initiation of cellular signals upon receptor ligation also depends on the association of cytoplasmic proteins. In fact, several proteins that can bind to TNF-R1 and TNF-R2 and are implicated in the signal transduction by these receptors have recently been identified (46–49). It is conceivable that the formation of such cytoplasmic signaling complexes is a time-dependent process and that the kinetic availability of such receptor-associated factors might govern the respective cellular sensitivity for the individual ligands.
Acknowledgments
We thank I.-M. von Broen (Knoll) for recombinant human TNF and G. Adolf (Bender MedSystems) for recombinant human LTα. This research was supported by the Deutsche Forschungsgemeinschaft (Gr 1307/3-1 and Gr 1307/3-2).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TNF, tumor necrosis factor; TNF-R, TNF receptor; LT, lymphotoxin; 125I-TNF, 125I-labeled TNF.
References
- 1.Damas P, Reuter A, Gysen P, Demonty J, Lamy M, Franchimont P. Crit Care Med. 1989;17:975–978. doi: 10.1097/00003246-198910000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Tracey K J, Cerami A. Annu Rev Cell Biol. 1993;9:317–343. doi: 10.1146/annurev.cb.09.110193.001533. [DOI] [PubMed] [Google Scholar]
- 3.Coffman F D, Green L M, Ware C F. Lymphokine Res. 1988;7:371–383. [PubMed] [Google Scholar]
- 4.Meager A, Sampson L E, Grell M, Scheurich P. Cytokine. 1993;5:556–563. doi: 10.1016/s1043-4666(05)80004-3. [DOI] [PubMed] [Google Scholar]
- 5.Chan H, Aggarwal B B. J Biol Chem. 1994;269:31424–31429. [PubMed] [Google Scholar]
- 6.Khabar K S, Siddiqui S, Armstrong J A. Immunol Lett. 1995;46:107–110. doi: 10.1016/0165-2478(95)00026-2. [DOI] [PubMed] [Google Scholar]
- 7.Smith C A, Farrah T, Goodwin R G. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 8.Tsujimoto M, Yip Y K, Vilcek J. Proc Natl Acad Sci USA. 1985;82:7626–7630. doi: 10.1073/pnas.82.22.7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kull F C, Jr, Jacobs S, Cuatrecasas P. Proc Natl Acad Sci USA. 1985;82:5756–5760. doi: 10.1073/pnas.82.17.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheurich P, Ücer U, Krönke M, Pfizenmaier K. Int J Cancer. 1986;38:127–133. doi: 10.1002/ijc.2910380120. [DOI] [PubMed] [Google Scholar]
- 11.Bajzer Z, Myers A C, Vuk-Pavlovic S. J Biol Chem. 1989;264:13623–13631. [PubMed] [Google Scholar]
- 12.Schall T J, Lewis M, Koller K J, Lee A, Rice G C, Wong G H W, Gatanaga T, Granger G A, Lentz R, Raab H, Kohr W J, Goeddel D V. Cell. 1990;61:361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- 13.Smith C A, Davis T, Anderson D, Solam L, Beckmann M P, Jerzy R, Dower S K, Cosman D, Goodwin R G. Science. 1990;258:1019–1023. doi: 10.1126/science.2160731. [DOI] [PubMed] [Google Scholar]
- 14.Hohmann H P, Brockhaus M, Baeuerle P A, Remy R, Kolbeck R, van Loon A P G M. J Biol Chem. 1990;265:22409–22417. [PubMed] [Google Scholar]
- 15.Pennica D, Lam V T, Mize N K, Weber R F, Lewis M, Fendly B M, Lipari M T, Goeddel D V. J Biol Chem. 1992;267:21172–21178. [PubMed] [Google Scholar]
- 16.Loetscher H, Stueber D, Banner D, Mackay F, Lesslauer W. J Biol Chem. 1993;268:26350–26357. [PubMed] [Google Scholar]
- 17.Grell M, Scheurich P, Meager A, Pfizenmaier K. Lymphokine Cytokine Res. 1993;12:143–148. [PubMed] [Google Scholar]
- 18.Moosmayer D, Dinkel A, Gerlach E, Hessabi B, Grell M, Pfizenmaier K, Scheurich P. Lymphokine Cytokine Res. 1994;13:295–301. [PubMed] [Google Scholar]
- 19.Thoma B, Grell M, Pfizenmaier K, Scheurich P. J Exp Med. 1990;172:1019–1023. doi: 10.1084/jem.172.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockhaus M, Schoenfeld H J, Schlaeger E J, Hunziker W, Lesslauer W, Loetscher H. Proc Natl Acad Sci USA. 1990;87:3127–3131. doi: 10.1073/pnas.87.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiers W. FEBS Lett. 1991;285:199–212. doi: 10.1016/0014-5793(91)80803-b. [DOI] [PubMed] [Google Scholar]
- 22.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Scheurich P. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 23.Tartaglia L A, Pennica D, Goeddel D V. J Biol Chem. 1993;268:18542–18548. [PubMed] [Google Scholar]
- 24.Hohmann H P, Remy R, Poschl B, van Loon A P. J Biol Chem. 1990;265:15183–15188. [PubMed] [Google Scholar]
- 25.Meager A, Leung H, Woolley J. J Immunol Methods. 1989;116:1–17. doi: 10.1016/0022-1759(89)90306-2. [DOI] [PubMed] [Google Scholar]
- 26.Old L J, Boyse E A, Clarke D A, Carswell E. Ann NY Acad Sci. 1962;101:80–106. [Google Scholar]
- 27.Dower S K, Titus J A, DeLisi C, Segal D M. Biochemistry. 1981;20:6335–6340. doi: 10.1021/bi00525a008. [DOI] [PubMed] [Google Scholar]
- 28.Higuchi M, Aggarwal B B. J Biol Chem. 1992;267:20892–20899. [PubMed] [Google Scholar]
- 29.Medvedev A E, Sundan A, Espevik T. Eur J Immunol. 1994;24:2842–2849. doi: 10.1002/eji.1830241139. [DOI] [PubMed] [Google Scholar]
- 30.Broudy V C, Harlan J M, Adamson J W. J Immunol. 1987;138:4298–4302. [PubMed] [Google Scholar]
- 31.Browning J, Ribolini A. J Immunol. 1989;143:1859–1867. [PubMed] [Google Scholar]
- 32.Ware C F, VanArsdale T L, Crowe P D, Browning J L. Curr Top Microbiol Immunol. 1995;198:175–218. doi: 10.1007/978-3-642-79414-8_11. [DOI] [PubMed] [Google Scholar]
- 33.Andrews J S, Berger A E, Ware C F. J Immunol. 1990;144:2582–2591. [PubMed] [Google Scholar]
- 34.Schuchmann M, Hess S, Bufler P, Brakebusch C, Wallach D, Porter A, Riethmuller G, Engelmann H. Eur J Immunol. 1995;25:2183–2189. doi: 10.1002/eji.1830250810. [DOI] [PubMed] [Google Scholar]
- 35.Grell M. J Inflamm. 1996;42:8–17. [PubMed] [Google Scholar]
- 36.Tartaglia L A, Goeddel D V. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 37.Pinckard J K, Sheehan K C F, Schreiber R D. J Biol Chem. 1997;272:10784–10789. doi: 10.1074/jbc.272.16.10784. [DOI] [PubMed] [Google Scholar]
- 38.Higuchi M, Aggarwal B B. J Immunol. 1994;152:3550–3558. [PubMed] [Google Scholar]
- 39.Porteu F, Hieblot C. J Biol Chem. 1994;269:2834–2840. [PubMed] [Google Scholar]
- 40.Evans T J, Moyes D, Carpenter A, Martin R, Loetscher H, Lesslauer W, Cohen J. J Exp Med. 1994;180:2173–2179. doi: 10.1084/jem.180.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshie O, Tada K, Ishida N. J Biochem. 1986;100:531–541. doi: 10.1093/oxfordjournals.jbchem.a121744. [DOI] [PubMed] [Google Scholar]
- 42.Bazzoni F, Beutler B. N Engl J Med. 1996;334:1717–1725. doi: 10.1056/NEJM199606273342607. [DOI] [PubMed] [Google Scholar]
- 43.Banner D W, D’Arcy A, Janes W, Gentz R, Schoenfeld H, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 44.Naismith J H, Devine T Q, Brandhuber B J, Sprang S R. J Biol Chem. 1995;270:13303–13307. doi: 10.1074/jbc.270.22.13303. [DOI] [PubMed] [Google Scholar]
- 45.Kischkel F C, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 47.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 48.Hsu H Y, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 49.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]