Abstract
Human protective protein/cathepsin A (PPCA), a serine carboxypeptidase, forms a multienzyme complex with β-galactosidase and neuraminidase and is required for the intralysosomal activity and stability of these two glycosidases. Genetic lesions in PPCA lead to a deficiency of β-galactosidase and neuraminidase that is manifest as the autosomal recessive lysosomal storage disorder galactosialidosis. Eleven amino acid substitutions identified in mutant PPCAs from clinically different galactosialidosis patients have now been modeled in the three-dimensional structure of the wild-type enzyme. Of these substitutions, 9 are located in positions likely to alter drastically the folding and stability of the variant protein. In contrast, the other 2 mutations that are associated with a more moderate clinical outcome and are characterized by residual mature protein appeared to have a milder effect on protein structure. Remarkably, none of the mutations occurred in the active site or at the protein surface, which would have disrupted the catalytic activity or protective function. Instead, analysis of the 11 mutations revealed a substantive correlation between the effect of the amino acid substitution on the integrity of protein structure and the general severity of the clinical phenotype. The high incidence of PPCA folding mutants in galactosialidosis reflects the fact that a single point mutation is unlikely to affect both the β-galactosidase and the neuraminidase binding sites of PPCA at the same time to produce the double glycosidase deficiency. Mutations in PPCA that result in defective folding, however, disrupt every function of PPCA simultaneously.
Galactosialidosis is a neurodegenerative lysosomal storage disease that is inherited as an autosomal recessive trait (1, 2). Patients are classified according to age of onset and the severity of symptoms as severe early infantile or milder late infantile and juvenile/adult. The clinical features include cardiac and kidney involvement, skeletal dysplasia, dysmorphism, angiokeratoma, progressive neurological deterioration, reduced life expectancy, and in some cases, mental retardation. At the biochemical level, galactosialidosis is diagnosed as a combined deficiency of β-galactosidase and neuraminidase activities in the lysosomes (3–5) that leads to storage of sialyloligosaccharides and glycopeptides in patient’s tissues and body fluids (ref. 2 and references therein). The primary defect, however, stems from the genetic alteration of a third lysosomal protein, protective protein/cathepsin A (PPCA) (1, 5).
PPCA is a multifunctional enzyme with distinct protective and catalytic activities (6). It is synthesized as a 452-amino acid (54 kDa) precursor (5), containing four disulfide bridges and two glycosylation sites (5, 7). In the endosomal/lysosomal compartment, the precursor undergoes endoproteolytic removal of a 2-kDa peptide, generating the mature and active form (5, 6, 8). The enzyme is active at both acidic and neutral pH and functions as cathepsin A/deamidase/esterase on a subset of neuropeptides (9–11). The dual and separable activities of the protein may reflect its capacity to participate in a variety of cellular processes not necessarily restricted to the lysosomal compartment, although its physiologic role is so far unknown. PPCA’s protective function resides in its ability to form a multienzyme complex with β-galactosidase and neuraminidase, contributing to the stability and lysosomal activity of both glycosidases (12, 13). Thus, the clinical phenotype of galactosialidosis is apparently dictated by the loss of PPCA protective function, with so far no discernible contribution from the cathepsin A deficiency (2). The interaction with PPCA is especially crucial for neuraminidase, which looses its enzymatic activity in absence of PPCA, whereas β-galactosidase maintains at least 10–15% of the normal enzyme values (2). In turn, the severe early-infantile forms of galactosialidosis present with features that are also observed in both the infantile and congenital type II sialidosis, caused by a single neuraminidase deficiency, and the GM1-gangliosidosis, caused by a primary defect in β-galactosidase (14, 15). This is consistent with PPCA deficiencies causing both glycosidases to malfunction.
The majority of the mutations identified so far in galactosialidosis patients are missense mutations, and as seen in other lysosomal storage disorders, the patient’s clinical severity often depends on the combination of the two mutant alleles encoding different PPCA variants (16–19). Many of these mutations were characterized according to their expression levels and functional deficits (16–18). For a few of them, homology modeling has been attempted by using the distantly related wheat serine carboxypeptidase structure (about 30% amino acid sequence identity) (20, 21). However, a precise structural understanding of how any of these mutations result in a dysfunctional protein can only come from the three-dimensional structure of PPCA itself (7). Herein, we have modeled 11 amino acid substitutions in the three-dimensional structure of the wild-type PPCA precursor. The results of this study give insights into the structural basis of this unusual lysosomal disease.
MATERIALS AND METHODS
Modeling studies of the mutations were performed by using the macromolecular modeling package O (22) on a Silicon Graphics interactive computer graphics system, Indigo XZ R4000. Further analysis of the effects of each amino acid substitution, including interatomic contacts and solvent-accessible surface calculations, was carried out by using programs from the ccp4 suite (23). The atomic structure used for this analysis has been refined to 2.2 Å with a crystallographic Rfactor of 21.3% (Rfree = 26.8%) (7). Figures were drawn with molscript (24) and raster3d (25).
RESULTS
We divided the 11 amino acid substitutions identified in PPCA variants from different galactosialidosis patients into two groups according to the ability of the mutant proteins to reach the lysosomes. Nine of the variants (17, 18) do not compartmentalize in lysosomes and have the following amino acid substitutions: Q21R, S23Y, W37R, S62L, V104M, L208P, Y367C, M378T, and G411S (group 1). Mutations Y221N and F412V result in a diminished but significant amount of protein still detectable in the lysosomes (group 2) (16, 18). A 12th mutation (SpDEx7) that generates an alternative splice site is leaky and allows for a small amount of correctly spliced mRNA resulting in low levels of wild-type protein (17). The mutations S23Y, S62L, V104M, L208P, Y367C, and G411S occur in compound heterozygosity with a second mutation from group 1 and are associated with a severe clinical manifestation of the disease (17, 18). In contrast, Y221N, F412V, SpDEx7, or a combination of these mutations results in a much milder phenotype (16–18). The mutations Q21R, W37R, and M378T have only been found in combination with F412V or SpDEx7. In these cases, a mild phenotype is observed that is likely dictated by the latter mutation (17, 18).
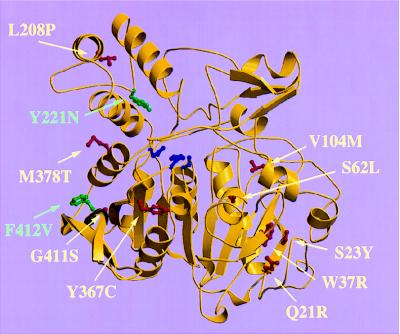
All 11 mutations were mapped in the three-dimensional structure of wild-type PPCA, and their effect on the structural integrity of the protein was assessed by substituting the wild-type residue in the atomic model of the normal PPCA precursor with the residue found in the variant. Nine mutations were localized in the core domain, whereas the other two were situated in the cap domain (Fig. 1).
Figure 1.
Schematic diagram of the PPCA monomer, which is present as a dimer in the crystal structure. The core domain contains the catalytic triad (shown in blue). The cap domain consists of a three-helical bundle and a small mixed β-sheet involved in enzyme inactivation. The PPCA mutations found in galactosialidosis patients are shown in red (group 1) or green (group 2).
Group 1.
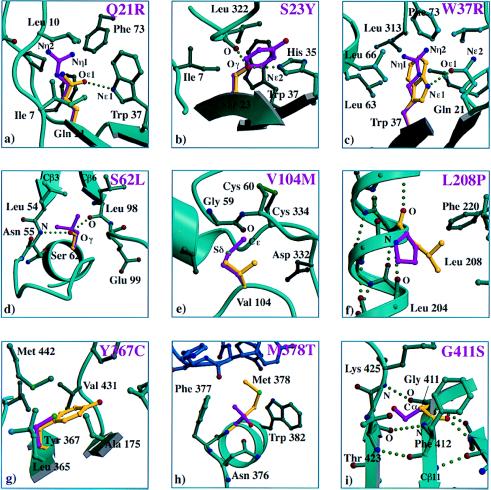
A panel of mutations from group 1 is shown in Fig. 2. The Q21R, S23Y, S62L, V104M, and G411S variants introduce a much bulkier side chain in the protein interior that would be predicted to cause severe steric clashes and to thereby have a detrimental effect on protein stability. For Q21R, S23Y, W37R, S62L, and L208P, hydrogen bond donors or acceptors are buried in the protein interior with no possibility for a partner. Loss of a hydrogen bonding partner is probably particularly detrimental for L208P, where a proline residue in the middle of helix Hα2 disrupts the hydrogen bonding pattern (Fig. 2f). The mutations Q21R and W37R introduce a positively charged residue in the interior of the protein with no apparent possibility of charge compensation, thereby decreasing the stability of these mutants. Internal cavities, formed by the introduction of smaller residues, might also contribute to protein destabilization for Y367C and L208P. For Y367C, introducing an extra free cysteine could have promoted the formation of improper disulfide bonds and concomitant misfolded intermediates. Although it is difficult to predict the precise effects of these mutations on protein structure without the crystal structure of the different mutant proteins, overall the impact of the group 1 mutations on protein folding is expected to be severe.
Figure 2.
Panel of PPCA mutations in group 1. Wild-type residues are shown in yellow, and the mutant side chains are shown in purple. Relevant side chains that interact with the residue of interest are depicted and labeled. β-strands, helices, and coils indicate the secondary structure elements that form the scaffold for the interacting residues. Color coding: carbon atoms, aqua spheres; nitrogen atoms, blue spheres; oxygen atoms, red spheres; sulfur atoms, green spheres; hydrogen bonds, green dotted lines. The impact of each mutation on the protein structure was assessed as follows. (a) Steric clashes due to incorporation of a larger side chain, positive charge introduced in hydrophobic interior, loss of hydrogen bond donor for Nɛ1 Trp37, no hydrogen bond acceptors for Nη1 Arg at position 21, and Nη2 Arg at position 21. (b) Steric clashes upon introduction of a larger side chain and loss of hydrogen bond to Nɛ2 His35. (c) Positive charge introduced in hydrophobic interior and no hydrogen bond acceptors for Nη1 Arg37 and Nη2 Arg37. (d) Steric clashes due to introduction of a larger side chain, forcing two β-strands (Cβ3 and Cβ6) in the central β-sheet apart, and loss of hydrogen bonds to main chain atoms: O Leu98 and N Asn55. (e) Steric clash with backbone carbonyl of Gly59 due to incorporation of a larger side chain while presence of disulfide Cys60–Cys334 limits possibilities for conformational changes accommodating the mutation. (f) Loss of hydrogen bond donor for main chain atom O Leu208, located in the middle of a helix, cavity created possibly disrupting packing. (g) Extra cysteine introduced, could promote formation of incorrect disulfides, smaller side chain incorporated introducing cavity. (h) New N-linked glycosylation site created at the dimer interface, and cavity introduced at the dimer interface due to incorporation of a smaller side chain. (i) Steric clashes due to incorporation of a larger side chain while backbone conformation at position 411 demands Ramachandran angles only favorable for Gly.
Similarly to Y367C, mutation M378T was modeled in the native structure without causing any strain because the substituted residue is smaller than the wild-type amino acid. However, it was shown that no functional protein is produced (18). The M378T substitution is peculiar in that it results in a small cavity centrally located at the dimer interface. More importantly, a consensus sequence for N-linked glycosylation was created by this substitution (Asn376-Xaa-Thr378). Given that biochemical studies have demonstrated that the M378T variant is more heavily glycosylated than the wild-type, these data strongly suggest that the additional glycosylation site is indeed used (18). Glycosylation of Asn376, also located at the dimer interface, must hinder dimerization and thereby cause retention of the variant protein in the endoplasmic reticulum (ER). The mutant W37R is puzzling in that biochemical studies would suggest a folded variant, because the protein is secreted; yet it is not detected in the lysosomes (17). The mutant side chain easily packs in the core of PPCA (see Fig. 2c) but introduces a positive charge in the protein interior without the possibility of charge compensation. This creates unsatisfied hydrogen bonds that probably destabilize the protein to such an extent that the variant protein either fails to reach the lysosomes or is degraded immediately in the organelle.
Three of the mutations were adjacent to each other in the three-dimensional structure: Q21R, S23Y, and W37R (Fig. 1). Ser23 and Trp37 are completely buried, whereas Gln21 is 98% buried with the Cγ and Nɛ2 atoms exposing only 3.7 Å2 of accessible surface to the solvent. Gln21 and Ser23 pack against opposite sides of Trp37, forming van der Waals contacts with Trp37 via side-chain carbon atoms. In addition, the Oɛ1 of Gln21 is hydrogen bonded to the Nɛ1 of Trp37 at an N–O distance of 3.0 Å. This surprising spatial concentration of residues, alteration of which has clear detrimental effects, may define a structurally or functionally sensitive region of the protein.
On the basis of the atomic model of normal PPCA, eight substitutions in group 1 are likely to have dramatic destabilizing effects on the protein monomer, whereas the ninth mutation affects the protein dimer. For the group 1 substitutions, the mean thermal B factors in the wild-type structure for residues Gln21, Ser23, Trp37, Ser62, Val104, Tyr367, and Gly411 fall between 7 and 13 Å2 (averaged over all atoms). This is lower than the average for all atoms in the dimer (17 Å2), as would be expected for core residues. The structural importance of residues Ser23, Trp37, Ser62, Val104, Leu208, Tyr367, and Gly411 in maintaining the protein fold is supported by the fact that they are almost completely conserved in the human, mouse, and chicken PPCAs, as well as in wheat and yeast serine carboxypeptidases (20, 26, 27). Consequently, the group 1 mutations probably destabilize the protein to such an extent that folding is severely hindered and the stability of the protein is drastically diminished. The misfolded mutant is retained to a large extent in the ER and essentially no functional protein can reach the lysosomes.
Group 2.
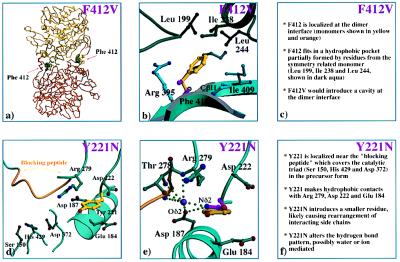
The mutations F412V and Y221N appear to have a markedly milder effect on the structural integrity of the protein compared with the group 1 mutations (Fig. 3). Phe412 is located in the core domain on strand Cβ11 at the dimer interface and is strictly conserved among the human, mouse, and chicken PPCAs (Figs. 1 and 3 a–c) (26). In the monomer, 68 Å2 of Phe412 is solvent-accessible, which equates to about 28% of its surface. Upon dimer formation, 83% of this exposed area (57 Å2) becomes buried through contacts with hydrophobic residues of the symmetry related monomer. As a result, the side chain of Phe412 is virtually inaccessible in the dimer with only 11 Å2 exposed to the solvent. Introducing a Val at position 412 creates a hydrophobic cavity of about 54 Å3 (calculated by using side-chain volumes given in ref. 28), which is expected to carry an unfavorable energetic price (Fig. 3b). Destabilization of the dimer interface at two places, one cavity at the surface of each subunit, would explain the biochemical observation that this variant is found largely as a monomer and is more rapidly degraded in the lysosome (16). Liao et al. (20) first suggested the importance of a large hydrophobic residue at position 412 for the dimer interface, pointing out that the dimeric wheat serine carboxypeptidase has a tryptophan at the equivalent position. Consistent with this argument, the yeast serine carboxypeptidase (27), which occurs only as a monomer, has a very different residue at the equivalent position, glutamic acid.
Figure 3.
Panel of PPCA mutants in group 2. (a) The locations of Phe412 in the PPCA precusor dimer. (b) Substitution of Phe412 (in yellow) with Val (in magenta). (c) The predicted effects of the F412V mutation on protein structural integrity. (d) The environment of Tyr221 in the PPCA dimer. (e) Substitution of Tyr221 (in yellow) with Asn (in magenta). A lilac sphere has been modeled to show how a water molecule or ion could participate in the interactions between Asn221 and the other side chains, allowing the formation of a more extensive hydrogen bonding network between the loop with Asn221, the blocking peptide, and the helix containing Asp187. (f) Proposed effects of the Y221N mutation on the local protein environment. Color coding: carbon atoms, aqua spheres; nitrogen atoms, blue spheres; oxygen atoms, red spheres; sulfur atoms, green spheres; blocking peptide (residues 272–277), orange; hydrogen bonds, green dotted lines.
The Y221N mutation appears both clinically and biochemically to be the mildest mutation known (18). Biochemical data have shown that only a fraction of the Y221N mutants reach the lysosome (16, 17) and that the serine carboxypeptidase activity is more drastically reduced than is the protective function (as monitored by the amount of β-galactosidase, neuraminidase, and cathepsin A activities still present) (17). Tyr221 is located in the cap domain, in a loop between Hα2 and Hα3 (Fig. 1), and is directly adjacent to Thr278 and Arg279 (Fig. 3 d–f). These residues are located C-terminally to the “blocking peptide” (residues 272–277), which fills the active site in the PPCA precursor rendering the enzyme catalytically inactive (7). The aromatic side chain of Tyr221 stacks between the aliphatic side chains of three surface residues [Glu184, Arg279, and Asp222 (Fig. 3 d and e)], forming extensive interactions with them. Carbon atoms of Glu184 (Cα, Cβ, Cγ, and Cδ), Asp222 (Cα, Cβ, and Cγ), and Arg279 (Cβ and Cγ) make good van der Waals contacts with atoms of the aromatic ring of Tyr221, with distances ranging from 3.5 to 4.5 Å. Although Tyr221 is near the surface, only 7% of the side chain, involving the hydroxyl group and the Cζ and the Cɛ2 atoms, is in contact with the solvent. An Asn residue can easily be incorporated at position 221 without causing steric clashes; however, this would drastically alter the hydrogen bonding network (Fig. 3e). It is more likely that the surrounding side chains rearrange to accommodate the smaller residue. Unlike its human and mouse counterparts, chicken PPCA has a His residue located at the position equivalent to Tyr221 (6). Comparison of the Y221N and chicken PPCA models shows that the Asn Nδ2 atom of the Y221N PPCA variant can be placed at a position corresponding to that of the Nδ1 atom of the imidazole ring of the chicken protein, but the Oδ1 atom of the Asn coincides with the Cδ2 of the imidazole ring (data not shown). This finding suggests that the Y221N mutation can be tolerated structurally to some extent. It is tempting to speculate that small rearrangements take place that alter the binding of the blocking peptide to the active site cleft in the Y221N precursor. In fact, in vitro enzyme activation of recombinant Y221N precursor with trypsin appears to only nick the protein and to not remove the 2-kDa excision peptide, as occurs in the wild-type enzyme (18). In this mutant, the enzymatic function is more likely to be impaired than is the protective function. However, biochemical studies have shown that the amount of Y221N enzyme detected in the lysosome is reduced (18). The primary effect of the Y221N amino acid substitution must, therefore, be on the efficiency of entry into the lysosome and on the stability of the variant in the lysosome.
Overall, molecular modeling of the Y221N and F412V substitutions in the three-dimensional structure of PPCA suggests that these mutations have a milder effect on protein structure than do the group 1 mutations. This is consistent with the biochemical observation that a small amount of group 2 mutant proteins still reaches the lysosomes and its residual activity causes a less-severe phenotype.
DISCUSSION
The localization of 11 naturally occurring PPCA mutations in the three-dimensional structure of the wild-type PPCA precursor is striking. None of the mutations is found directly at the protein surface or in the active site, indicating that none of them brings about its detrimental effects by directly disrupting the “protective function” or the “catalytic machinery” of PPCA. This is atypical among the numerous genetic deficiencies for which a three-dimensional structure of the affected protein is known or for which a homology model is available. These include hemoglobin (29), apolipoprotein E (30, 31), factor IX (32–34), antithrombin III (35), α1-antitrypsin (36), medium-chain acyl-CoA dehydrogenase (37, 38), and myosin (39). Mutations in globular proteins that are associated with genetic diseases are usually found to be distributed into different categories according to how they predominantly affect the function or structure of the protein. Major categories are (i) impairment of the catalytic machinery; (ii) disruption of a functional site, such as substrate or cofactor binding sites, receptor binding regions, or protein processing loops; (iii) modification of intersubunit interfaces, thereby preventing correct oligomerization or creating undesirable aggregation states; and (iv) destabilization of the protein.
All of the mutations in human PPCA that we analyzed, with the exception of F412V and Y221N, disrupted properties necessary for correct protein folding (40, 41). This predominance of folding defects as the molecular basis for galactosialidosis is likely shared by other lysosomal enzyme deficiencies. The folding defects appeared to result from (i) introducing unsatisfied charged groups and hydrogen bonds donors/acceptors to the protein core; (ii) introducing steric clashes, because of a bulkier side chain, that disrupt the packing of the protein core; and (iii) creating cavities in protein interiors and interfaces. Such defects in proteins that are normally transported through the ER and Golgi apparatus, such as plasma membrane or lysosomal proteins, are likely to have a profound effect on their localization. It is now clear that cells have developed quality-control mechanisms that retain proteins that are not correctly folded or assembled after synthesis, for refolding or degradation (42, 43). The efficient release of normal PPCA precursor from the ER en route to the lysosome depends on its early oligomerization, which probably requires accurate folding (16). Coupled to these two events is the interaction of the precursor with neuraminidase and β-galactosidase that is known to occur shortly after synthesis and to influence the intracellular targeting of the two glycosidases (ref. 44 and A.d.A. unpublished results). Thus, a genetic lesion that would affect proper folding of the precursor could impair all other functions of the protein from synthesis on and could ultimately influence the fate of neuraminidase and β-galactosidase, as would null mutants of PPCA. This would explain why virtually all of the protective protein variants discovered so far in galactosialidosis patients appear to affect folding. If the binding sites for β-galactosidase and neuraminidase are distinct on the protective protein, complete loss of the multienzyme complex would require at least two drastic mutations per allele to disrupt both binding sites. The probability of such an event is extremely low. However, removal of the protective protein (in part or totally) through the introduction of folding defects could achieve the same effect with a single mutation per allele. Can we predict the identification of patients with mutations that do disrupt the binding sites for either neuraminidase or β-galactosidase? Such patients might be difficult to identify, because they may be clinically and biochemically indistinguishable from classic sialidosis or GM1-gangliosidosis, and they may have completely normal cathepsin A activity. It is, therefore, conceivable that patients presenting clinically with a sialidosis phenotype may not carry a mutation in the neuraminidase structural gene but instead may carry a mutation in the PPCA binding site for neuraminidase (45).
The F412V mutation raises an interesting point because it is located at the dimer interface (this paper) and was shown to impair proper dimerization of the precursor (16). It is possible that the extent to which heterodimers can be formed between different PPCA variants or mutant and wild-type polypeptides will influence the amount of PPCA retained or discharged from the ER. It is unknown whether oligomerization occurs before, during, or after folding of the PPCA precursor. However, one could envisage that a “milder” mutation such as F412V, when present in heterozygosity with the product of the normal allele, could still be able to partially interact with the normal protein, thereby depleting the total pool of normal molecules reaching the lysosomes. The end effect on the two glycosidases, particularly neuraminidase, would be that of a dominant negative mutation. In line with this assumption, we have noticed that in the parents of a late-infantile galactosialidosis, homozygous for the F412V mutation, the lysosomal activity of neuraminidase was exceptionally low, although they never developed the disease (46).
The data presented herein provide a structural explanation for the failure of some mutant PPCAs to reach the lysosomes and further substantiate the observed correlation between the clinical severity of galactosialidosis and the amount of residual protein found in lysosomes.
Acknowledgments
We thank Christophe Verlinde and Ethan Merritt for managing the computer facilities at the Biomolecular Structure Center. Support from Prof. H. Galjaard and the Clinical Genetics Foundation Rotterdam in the initial stages of the structure determination has been invaluable. W.G.J.H. acknowledges receipt of a major equipment grant from the Murdock Charitable Trust and the support of the University of Washington School of Medicine. These studies were supported, in part, by the National Institutes of Health Cancer Center Support (CORE) Grant P30-CA21765 and by the American Lebanese Syrian Associated Charities.
ABBREVIATIONS
- PPCA
protective protein/cathepsin A
- ER
endoplasmic reticulum
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1IVY).
References
- 1.D’ Azzo A, Hoogeveen A, Reuser A J J, Robinson D, Galjaard H. Proc Natl Acad Sci USA. 1982;79:4535–4539. doi: 10.1073/pnas.79.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.d’Azzo A, Andria G, Strisciuglio P, Galjaard H. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2825–2837. [Google Scholar]
- 3.Wenger D A, Tarby T J, Wharton C. Biochem Biophys Res Commun. 1978;82:589–595. doi: 10.1016/0006-291x(78)90915-4. [DOI] [PubMed] [Google Scholar]
- 4.Andria G, Strisciuglio P, Pontarelli G, Sly W S, Dodson W E. In: Sialidases and Sialidosis. Perspectives in Inherited Metabolic Diseases. Tettamanti G, Durand P, Di Donato S, editors. Milano, Italy: Edizioni Ermes; 1981. pp. 379–395. [Google Scholar]
- 5.Galjart N J, Gillemans N, Harris A, van der Horst G T J, Verheijen F W, Galjaard H, d’Azzo A. Cell. 1988;54:755–764. doi: 10.1016/s0092-8674(88)90999-3. [DOI] [PubMed] [Google Scholar]
- 6.Galjart N J, Morreau H, Willemsen R, Gillemans N, Bonten E J, d’Azzo A. J Biol Chem. 1991;266:14754–14762. [PubMed] [Google Scholar]
- 7.Rudenko G, Bonten E J, d’Azzo A, Hol W G J. Structure. 1995;3:1249–1259. doi: 10.1016/s0969-2126(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 8.Bonten E J, Galjart N J, Willemsen R, Usmany M, Vlak J M, d’Azzo A. J Biol Chem. 1995;270:26441–26445. doi: 10.1074/jbc.270.44.26441. [DOI] [PubMed] [Google Scholar]
- 9.Jackman H L, Tan F L, Tamei H, Buerling-Harbury C, Li X Y, Skidgel R A, Erdos E G. J Biol Chem. 1990;265:11265–11272. [PubMed] [Google Scholar]
- 10.Jackman H L, Morris P W, Deddish P A, Skidgel R A, Erdos E G. J Biol Chem. 1992;267:2872–2875. [PubMed] [Google Scholar]
- 11.Hanna W L, Turbov J M, Jackman H L, Tan F, Froelich C J. J Immunol. 1994;153:4663–4672. [PubMed] [Google Scholar]
- 12.Verheijen F, Brossmer R, Galjaard H. Biochem Biophys Res Commun. 1982;108:868–875. doi: 10.1016/0006-291x(82)90911-1. [DOI] [PubMed] [Google Scholar]
- 13.Verheijen F W, Palmeri S, Hoogeveen A T, Galjaard H. Eur J Biochem. 1985;149:315–321. doi: 10.1111/j.1432-1033.1985.tb08928.x. [DOI] [PubMed] [Google Scholar]
- 14.Thomas G H, Beaudet A L. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2529–2561. [Google Scholar]
- 15.Suzuki Y, Sakuraba H, Oshima A. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2785–2823. [Google Scholar]
- 16.Zhou X-Y, Galjart N J, Willemsen R, Gillemans N, Galjaard H, d’Azzo A. EMBO J. 1991;10:4041–4048. doi: 10.1002/j.1460-2075.1991.tb04980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimmoto M, Fukuhara Y, Itoh K, Oshima A, Sakuraba H, Suzuki Y. J Clin Invest. 1993;91:2393–2398. doi: 10.1172/JCI116472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X-Y, van der Spoel A, Rottier R, Hale G, Willemsen R, Berry G T, Strisciuglio P, Andria G, d’Azzo A. Hum Mol Genet. 1996;5:1977–1987. doi: 10.1093/hmg/5.12.1977. [DOI] [PubMed] [Google Scholar]
- 19.Neufeld E F. Annu Rev Biochem. 1991;60:257–280. doi: 10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- 20.Liao D-I, Breddam K, Sweet R M, Bullock T, Remington S J. Biochemistry. 1992;31:9796–9812. doi: 10.1021/bi00155a037. [DOI] [PubMed] [Google Scholar]
- 21.Elsliger M-A, Potier M. Proteins: Structure, Function and Genetics. 1994;18:81–93. doi: 10.1002/prot.340180110. [DOI] [PubMed] [Google Scholar]
- 22.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computing Project. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 24.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 25.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 26.Galjart N J, Gillemans N, Meijer D, d’ Azzo A. J Biol Chem. 1990;265:4678–4684. [PubMed] [Google Scholar]
- 27.Endrizzi J A, Breddam K, Remington S J. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- 28.Harpaz Y, Gerstein M, Chothia C. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 29.Williamson D. Blood Rev. 1993;7:146–163. doi: 10.1016/0268-960x(93)90002-l. [DOI] [PubMed] [Google Scholar]
- 30.Wilson C, Wardell M R, Weisgraber K H, Mahley R W, Agard D A. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 31.Wilson C, Mau T, Weisgraber K H, Wardell M R, Mahley R W, Agard D A. Structure. 1994;2:713–718. doi: 10.1016/s0969-2126(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 32.Tsang T C, Bentley D R, Mibashan R S, Giannelli F. EMBO J. 1988;7:3009–3015. doi: 10.1002/j.1460-2075.1988.tb03164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamaguchi N, Roberts H, Stafford D W. Biochemistry. 1993;32:6324–6329. doi: 10.1021/bi00076a004. [DOI] [PubMed] [Google Scholar]
- 34.Lin S-W, Lin C-N, Hamaguchi N, Smith K J, Shen M-C. Blood. 1994;84:1866–1873. [PubMed] [Google Scholar]
- 35.Cooper D N. Ballière’s Clinical Haemotology. 1994;7:637–674. doi: 10.1016/s0950-3536(05)80102-7. [DOI] [PubMed] [Google Scholar]
- 36.Stein P E, Carrell R W. Nat Struct Biol. 1995;2:96–113. doi: 10.1038/nsb0295-96. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka K, Yokota I, Coates P M, Strauss A W, Kelly D P, Zhang Z, Gregersen N, Andresen B S, Matsubara Y, Curtis D, Chen Y T. Hum Mutat. 1992;1:271–279. doi: 10.1002/humu.1380010402. [DOI] [PubMed] [Google Scholar]
- 38.Andresen B S, Jensen T G, Bross P, Knudsen I, Winter V, Kølvraa S, Bolund L, Ding J H, Chen Y T, van Hove J L K, Curtis D, Yokota I, Tanaka K, Park Kim J J, Gregersen N. Am J Hum Genet. 1994;54:975–988. [PMC free article] [PubMed] [Google Scholar]
- 39.Rayment I, Holden H, Sellers J R, Fananapazir L, Epstein N D. Proc Natl Acad Sci USA. 1995;92:3864–3868. doi: 10.1073/pnas.92.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews B W. Annu Rev Biochem. 1993;62:139–160. doi: 10.1146/annurev.bi.62.070193.001035. [DOI] [PubMed] [Google Scholar]
- 41.Matthews B W. Nat Struct Biol. 1995;2:85–86. doi: 10.1038/nsb0295-85. [DOI] [PubMed] [Google Scholar]
- 42.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 43.Gottesmann S, Wickner S, Maurizi M R. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 44.Morreau H, Galjart N J, Willemsen R, Gillemans N, Zhou X-Y, d’Azzo A. J Biol Chem. 1992;267:17949–17956. [PubMed] [Google Scholar]
- 45.Bonten E, van der Spoel A, Fornerod M, Grosveld G, d’Azzo A. Genes Dev. 1996;10:3156–3169. doi: 10.1101/gad.10.24.3156. [DOI] [PubMed] [Google Scholar]
- 46.Kleijer W J, Geilen G C, Janse H C, van Diggelen O P, Zhou X-Y, Galjart N J, Galjaard H, d’Azzo A. Pediatr Res. 1996;39:1067–1071. doi: 10.1203/00006450-199606000-00022. [DOI] [PubMed] [Google Scholar]