Abstract
Gene silencing in plants inactivates transgenes introduced into plants and/or endogenous homologous genes. This stable but potentially reversible loss of gene activity resembles epigenetic changes that occur in normal development. The stability of silencing implies the involvement of trans-acting components, although none of them have been identified so far. Here we report the finding of second-site mutations interfering with maintenance of the silent state. We mutagenized Arabidopsis thaliana plants carrying a silent transgene encoding hygromycin phosphotransferase (hpt) and therefore show a heritable hygromycin-sensitive phenotype. The M2 generation was screened for hygromycin resistance. Eight putative mutants (som1 to 8) were found that expressed the transgene and transmitted the expressed state to their progeny. All mutations were shown to reactivate a silent transgenic test locus in trans. The level of DNA methylation at the hpt locus and at centromeric repeats was found to be reduced in the som mutants. Complementation crosses indicated complex epigenetic interactions among the som mutant alleles and with the previously described ddm1 allele, which elicits DNA hypomethylation [Vongs, A., Kakutani, T., Martienssen, R. A. & Richards, E. J. (1993) Science 260, 1926–1928]. Som mutants can be classified into three groups: (i) allelic or interacting with ddm1 and with each other (som 1, 4, and 5), (ii) nonallelic with ddm1 and som mutants of group A (som2), and (iii) mutants with slow resilencing after outcrosses, which hinders their classification (som 3, 6, 7, and 8).
The loss of expression of previously active genes in plants, also referred to as gene silencing (1), is observed in response to developmental, environmental, or unknown signals. It occurs at a frequency higher than that of mutations, yet is markedly stable during somatic transmission. Gene silencing, initially perceived mainly as an unwanted source of instability of transgene expression, is now regarded as a molecular tool to intentionally regulate gene expression (2, 3).
In spite of increasing evidence for epigenetic gene silencing in many different systems, we are far from understanding its molecular basis. In all cases, gene silencing is specified by a drastically reduced steady-state level of mRNA. It appears that chromosomal position and/or structure of the affected loci are factors determining the frequency and strength of silencing. Inactivation seems to preferentially affect genes present in multiple copies and is thought to be a consequence of sequence redundancy. After the initial observation of heritable silencing in the case of two partially homologous transgenic loci (4), many other examples of homology-dependent gene silencing have been reported (for review, see refs. 2, 5, 6). Closer analysis has allowed the classification of silencing events according to the relative position of the affected loci (cis, trans, allelic, ectopic), the origin of the affected genes (endogenous and/or transgenic), and the level of interaction (posttranscriptional or transcriptional) (3). Posttranscriptional silencing seems mainly to involve the formation of aberrant RNA molecules and is occasionally, but not necessarily, accompanied by DNA methylation. Silencing that interferes with transcription initiation is more strictly correlated with hypermethylation of the DNA and possibly with alteration of chromatin structure at the silent loci (5, 6). It is, however, not clear whether these molecular events are a prerequisite for epigenetic silencing or a consequence of the silent state. The cellular functions involved in the switch from active to inactive genes are not known.
Epigenetic modification of gene expression is also well known in fungi (7, 8) and insects (9). Substantial insight into the regulatory mechanisms of gene silencing in yeast and Drosophila was obtained from studies of second-site modifier mutations interfering with the silencing process in trans (10, 11). In plants, the first mutant screen described was designed to isolate mutants with enhanced gene silencing activity (egs). Two lines of Arabidopsis with independent mutant loci egs1 and egs2 (12) were isolated from an M2 population by a direct screen for silencing of an Agrobacterium rhizogenes rolB transgene. When active, this gene visibly alters plant development. Recently, egs1 was shown to be a trans-acting mutation not connected with the rolB gene used for its selection: when egs1 was crossed into a line containing an active rolB transgene, F2 progeny included individuals with an inactive rolB locus (Sarah Grant, personal communication). Thus, the egs1 mutation appears to lead to the inactivation of this rolB transgene, and, consequently, the wild-type allele may actively prevent silencing.
Because in the case of transcriptional silencing the inactive state of affected genes is stably transmitted through mitotic and meiotic divisions, trans-acting modifier loci may be responsible for the stability of epigenetic suppression. Mutations in such loci can be predicted to cause reduced gene silencing, due to reactivation of previously silent loci by interference with maintenance of the silent state or by a failure to recognize sequence redundancy. Here we describe a selection for this type of mutation, based on mutagenesis of an Arabidopsis line carrying a heritably inactivated, methylated hygromycin-resistance gene. We report the identification of eight Arabidopsis lines with restored hygromycin resistance. These mutations have been proven to release silencing of a transgenic locus in trans. Results presented here document the involvement of cellular functions in the maintenance of transcriptional silencing during mitotic and meiotic divisions. The recovery of mutants further suggests that the level of epigenetic silencing can be modified without causing lethality or severe phenotypic changes.
MATERIALS AND METHODS
Mutagenesis.
The transgenic line A of Arabidopsis thaliana ecotype Zürich, with a stably silenced locus containing multiple copies of a chimeric hygromycin phosphotransferase gene (hpt), was described previously (13, 14). The original tetraploid line was bred into a diploid derivative by double backcross to the diploid wild type (wt). Among the progeny of a single plant hemizygous for the transgene, 100 plants containing the transgene were identified by PCR. They were expected to consist of one-third and two-thirds with a homo- or hemizygous configuration of the transgene, respectively. These plants yielded approximately 150,000 seeds. Ethylmethane sulfonate (EMS) mutagenesis was performed by incubating approximately 50,000 seeds (M1) in 0.3% EMS for 12 hr with gentle shaking, followed by a thorough washing with water. A control batch of 10,000 seeds was incubated for 12 hr in water. Fast neutron irradiation (FNR) mutagenesis was performed by exposing approximately 75,000 M1 seeds to FNR dose of 60 Gy. M1 plants were grown to maturity in trays of soil (27 cm × 44 cm) at 20°C, 70% relative humidity and a 12-hr/12-hr light/dark cycle. To minimize seed loss, seeds were harvested every 3 to 4 days by using a vacuum cleaner with a fine-mesh tea filter. M2 seeds were collected separately from each tray, resulting in 20 and 30 M2 families after EMS and FNR mutagenesis, respectively. Seeds of control plants were bulk-harvested.
Selection of Putative Mutants.
Before screening, M2 seeds were dried for 1 week at room temperature and cold-treated at 4°C for a minimum of 1 week. They were surface-sterilized twice with calcium hypochlorite (5%, with 0.1% Tween 80) for 15 min and washed with sterile double-distilled water. For selection, seeds were plated in aseptic conditions on germination medium (15) solidified with 0.8% agar and containing 15 mg/l hygromycin B (Calbiochem). Approximately 2,000 seeds were plated on 14-cm Petri dishes containing 100 ml medium. To ensure equal distribution during sowing, seeds were mixed with 30 ml of the same medium containing 0.4% agar. Two seeds from a hygromycin-resistant line were sown at marked locations on each plate as positive controls. Plates were incubated at 16 hr light/20°C and 8 hr dark/16°C and evaluated for hygromycin resistance each day for 8–15 days after sowing. Seedlings showing hygromycin resistance were transferred to larger vessels containing germination medium without hygromycin. After rosette formation and development of the root system, plants were transferred to soil for further growth and seed setting. Before potting, tissue explants were taken from each plant to generate callus cultures on MSPII medium (16) with or without 15 mg/l hygromycin B.
DNA and RNA Analysis.
Genomic DNA was isolated from callus material by using N-cetyl-N,N,N-trimethylammonium bromide (CTAB) as described (17). Conditions of Southern blot analysis were described previously in detail (13). Total RNA from callus material was obtained by using RNA easy columns (Qiagen) according to the supplier’s recommendation and analyzed by Northern blotting as described (13). PCR analysis was performed by using denatured leaf extracts (18). Primers specific for the hpt-coding region and the PCR conditions have been described (17). Primers amplifying a slightly larger fragment of the alcohol dehydrogenase gene (5′-GAG AAG GAG TGA CTG ATC TTC AGC and 5′-CAA CAC CGA TGA TCC TAG AAG CAC) were used as internal controls.
Genetic Crosses and Assay for Resistance.
The crosses were performed as described (13). For trans-activation crosses, line A homozygous for the silent hpt gene was used. Mature seeds were harvested approximately 3 weeks after crossing. Selfed or hybrid progeny seeds were surface-sterilized and grown on germination medium (0.8% agar) in 9-cm Petri dishes under the conditions described above, at the hygromycin concentrations indicated in the Results section.
RESULTS
Mutagenesis and Mutant Selection.
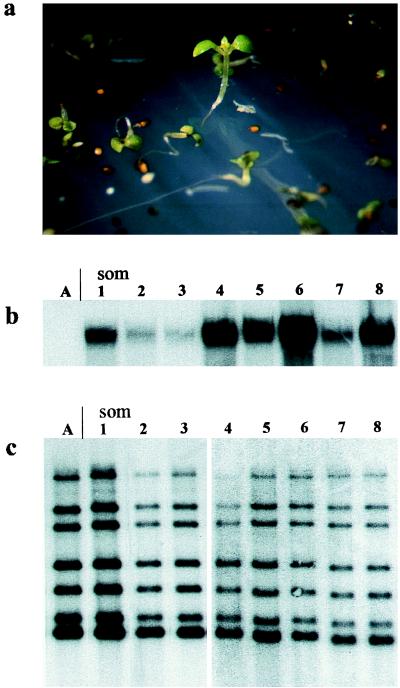
The identification of potential mutations that would interfere with silencing was based on two different mutagenic treatments of the transgenic Arabidopsis line. This line contained a locus with multiple copies of the hygromycin phosphotransferase gene, stably silenced over several generations and therefore sensitive to hygromycin (14). Release of silencing from this locus would render a hygromycin-resistant phenotype. Because no spontaneous reactivation was found in a large, nonmutagenized control population (74,000 seedlings), recovery of resistance was expected to originate only from mutagenesis. Treatment with EMS reduced the germination rate of the M1 seeds to 33%, whereas FNR resulted in 93% germination. The seed yield of the M1 populations was ca. 25%, compared with the mock-treated plants grown under the same conditions. The screening of 80,000 M2 seeds after EMS mutagenesis and 120,000 M2 seeds after FNR resulted in 17 and 43 plantlets, respectively, which survived for at least 8 days on hygromycin-containing medium (Fig. 1a).
Figure 1.
Selection of mutants. Primary selection of mutant candidates: hygromycin-resistant seedling after 8 days selection on 15 mg/l hygromycin (a), Northern (b), and Southern (c) blot analysis of the parental line A with the silent hpt gene and the mutants som1-som8. Both membranes were probed with the hpt coding region. Genomic DNA was digested with BamHI.
Hygromycin-resistant seedlings, recovered from bulk selection, were rescued and grown further on antibiotic-free medium. This allowed the recovery of individuals with only partial reactivation of the transgenic locus, but may also have resulted in the rescue of false positives. Thus, all individuals recovered from the primary selection were examined further for restoration of gene expression from the previously silent hpt gene by using three independent assays: (i) confirmation of a hygromycin-resistant phenotype in tissue culture, (ii) activity of the transgenic locus by mRNA analysis, and (iii) integrity of the transgenic locus determined by genomic DNA analysis.
Assay of hygromycin resistance on the basis of tissue culture response was performed on leaf explants taken from rescued plants after several weeks of growth on nonselective medium. Tissue from plants with a reactivated transgene was expected to form callus on hygromycin-containing medium, similar to the response without the antibiotic. However, hygromycin resistance could have resulted from a reduced uptake or a metabolic resistance to the antibiotic. Thus, replica callus tissue derived from explants grown under nonselective conditions was used as a source of RNA for Northern blot analysis (Fig. 1b). Only mutant candidates with a detectable level of hpt mRNA were studied further.
Restored expression of a previously silent multicopy transgenic locus might have been the result of recombination and reduction of the copy number (17, 19), perhaps induced by the mutagenic agents. To exclude such events, DNA from callus tissue of hygromycin-resistant clones was subjected to Southern blot analysis. All selected clones preserved the complex banding pattern specific for the transgenic insert of the parental line (Fig. 1c). There was no indication of recombination within the transgenic locus, and the hpt transcript was likely to be derived from the previously silent multicopy locus.
All three criteria for transcriptional reactivation of the silent locus were met by 5 and 21 M2 plants after EMS and FNR mutagenesis, respectively. These 26 putative mutants were derived from three and five independent M2 families after EMS and FNR treatment, respectively. They were termed som, after the somniferous effect of the wild-type allele on susceptible genes. One putative mutant from each M2 family was chosen for further studies (Table 1).
Table 1.
Isolation and characterization of som mutants
| Mutant
|
||||||||
|---|---|---|---|---|---|---|---|---|
| som1 | som2 | som3 | som4 | som5 | som6 | som7 | som8 | |
| Mutagen | EMS | EMS | EMS | FNR | FNR | FNR | FNR | FNR |
| Hygromycin resistance | + | + | + | + | + | + | + | + |
| Transcriptional activity | + | + | + | + | + | + | + | + |
| Integrity of transgenic insert | + | + | + | + | + | + | + | + |
| Other candidates in the same M2 family | 0 | 0 | 2 | 4 | 4 | 0 | 3 | 5 |
| Self-fertility | + | + | − | + | + | + | + | + |
| Transgene genotype | Hemizygous | Hemizygous | Homozygous | Homozygous | Homozygous | Homozygous | Hemizygous | Hemizygous |
| Resistance in selfed progeny | + | + | + | + | + | + | + | |
| Number of seedlings tested | 117 | 116 | 67 | 29 | 26 | 67 | 60 | |
| Number of resistant seedlings | 50 | 69 | 59 | 25 | 18 | 40 | 35 | |
| Percentage of resistance | 43% | 60% | 88% | 86% | 69% | 60% | 58% | |
| Trans-activation of silent locus* | + | + | + | + | + | + | + | + |
| Number of seedlings tested | 373 | 626 | 175 | 133 | 126 | 141 | 289 | 125 |
| Number of resistant seedlings | 45 | 98 | 27 | 26 | 18 | 32 | 45 | 19 |
| Percentage of resistance | 12.1% | 15.7% | 15.4% | 19.5% | 14.3% | 22.7% | 15.6% | 15.2% |
*Compare with Fig. 2.
Inheritance of Resistance.
Assuming single mutations, putative mutants with hemi- or homozygous transgenes were expected to yield 75 and 100% resistant plants, respectively, after self-pollination. Progeny of som1 and som2 (EMS) as well as som4 to som8 (FNR) mutant candidates were selected for hygromycin resistance (som3 was not self-fertile). In all cases, the restored resistance was genetically transmitted, although with significantly lower representation of resistant progeny for som1 and som6 (Table 1). This could be due to distortion of segregation by other mutations, or due to incomplete or unstable reactivation of the hpt locus. In the case of som1, a trans-acting mutation may have been able to reactivate only the segregating hemizygous configuration of the hpt locus.
Trans-Acting Effects of Mutant Loci.
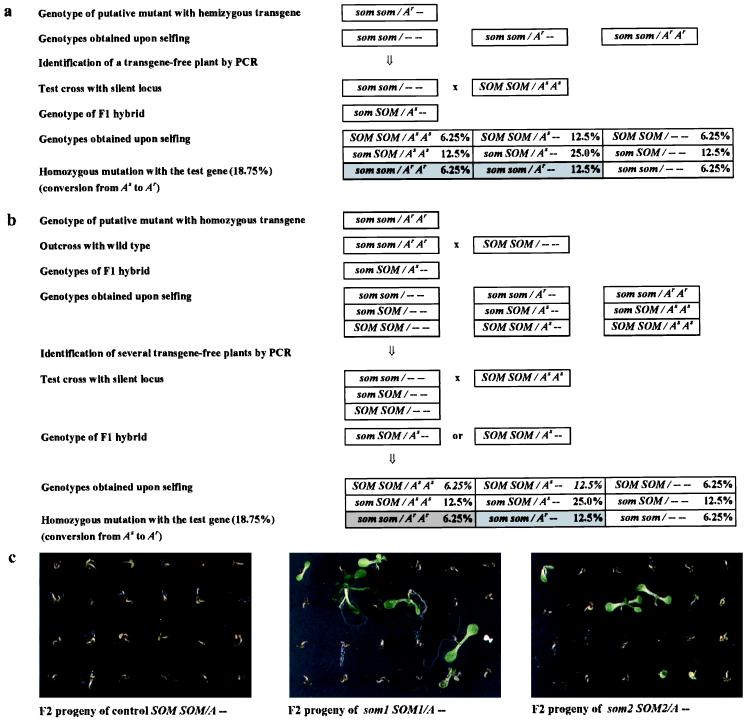
Restoration of gene activity and inheritance of hygromycin resistance could have originated either from cis-acting mutations within the transgene or from mutations reactivating the marker gene in trans. To distinguish between these possibilities, the putative som loci were separated from the residing transgene and retested on a new, silent allele. The mutant candidates with the hpt locus in hemizygous configuration allowed an easy separation of trans-acting som loci and the residing hpt gene by segregation in the M3 (Fig. 2a). Mutants with homozygous hpt locus were outcrossed to the wild type, and four F2 plants each derived from som 3, 4, 5, and 6, lacking the transgene, were identified, half of them expected to contain the mutated som alleles (Fig. 2b). In both cases, the transgene-free plants were crossed with plants homozygous for the stably inactive hpt gene as a test locus (Fig. 2). Segregating F2 populations of these crosses for every som mutant contained resistant seedlings (12% to 23%, Fig. 2c and Table 1). These values were compatible with the expected ratio of 18.75% and 12.5% for a recessive mutant genotype trans-activating the hemi- and homozygous, or only the hemizygous hpt gene, respectively (Fig. 2). Therefore, it can be concluded that all analyzed som mutations represent trans-acting traits able to release silencing from a previously inactive transgenic locus.
Figure 2.
Reactivation of a silent gene in test crosses with the mutants. (a) Scheme of the crosses for mutants with hemizygous transgene. (b) Scheme of the crosses for mutants with homozygous transgene. (c) F2 populations after 2 weeks on 10 mg/l hygromycin. som, mutant locus; SOM, corresponding wild-type allele; As, hpt gene in silent state; Ar, hpt gene in reactivated state; −, transgene-free locus allelic to A.
Analysis of DNA Methylation.
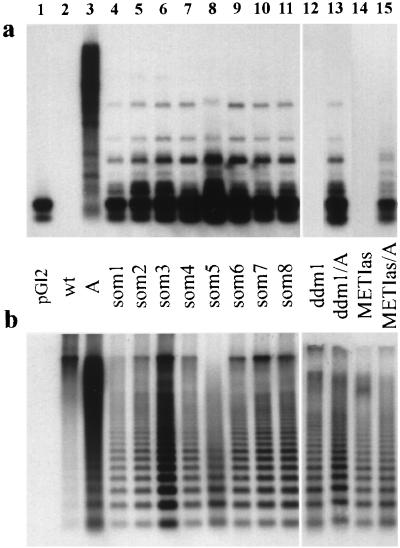
Because the silent transgene in line A was hypermethylated, analogous test crosses of A were performed with two strains characterized by genome-wide hypomethylation: the mutant ddm1 (20) and a line transgenic for a methyltransferase antisense gene (METIas; ref. 21). Resistant seedlings (13.6%) were recovered only among F2 progenies from the cross with ddm1, resembling the effect of the som mutations. Southern blot analysis with a methylation-sensitive restriction endonuclease revealed that, compared with the parental line A, all som mutants and the hygromycin-resistant F2 progeny of ddm1/A had a reduced methylation level at the hpt locus as well as at centromeric repeats (Fig. 3). Although DNA of the METIas/A progeny was hypomethylated to the same extent (Fig. 3), no hygromycin-resistant seedling could be recovered from this F2 population. Therefore, a reduced activity of methyltransferase is not sufficient to release silencing from the A locus and is probably not the basis of the som mutations. The data suggest that som mutations are resembling ddm1, being either epistatic or allelic.
Figure 3.
Methylation analysis. The same Southern blot of DNA cut with HpaII was probed with the hpt coding region (a), or with a fragment containing the centromeric 180-bp repeat (b; ref. 20). Lanes: 1, pGl2 plasmid used to generate the parental line A; 2, wt; 3, parental line A; 4–11, som 1–8, 12, mutant ddm1/ddm1 with reduced methylation (20); 13, ddm1/A; 14, METIas T3 #10.5 transgenic for a methyltransferase antisense construct (21); 15, METIas T3 #10.5/A.
Complementation Crosses.
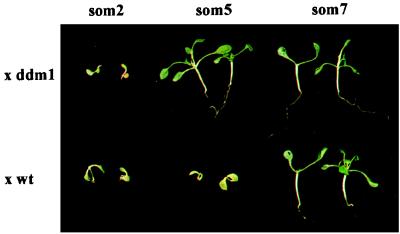
To detect a possible allelism between som and ddm1, as well as among the som mutations, crosses were performed between the som mutants containing the hpt locus and the ddm1 mutant or the wild type. If the mutations would be allelic, the F1 hybrids were expected to maintain the resistance to hygromycin, whereas a cross to nonallelic, recessive mutations or to the wild type could allow resilencing of the transgene, leading to a loss of resistance. However, the results were not as explicit as that due to lasting effects of epigenetic states inherited from the parental lines. Nevertheless, a cautious evaluation based on the F1 phenotypes allowed us to distinguish three groups (Fig. 4): class A with resistant som/ddm1, sensitive som/wt hybrids, and resistant hybrids among each other (som1, 4 and 5), class B with sensitive som/ddm1, sensitive som/wt hybrids, and also sensitive after crosses with som mutants of group A. This group contained only one member (som2). Class C includes mutants with only resistant hybrids (som3, 6, 7, and 8), indicating a pronounced delay of resilencing of the hpt gene, which obstructed the determination of their true genetic or epigenetic interactions with other alleles.
Figure 4.
Complementation analysis. F1 hybrids of som2, som5, and som7 with ddm1 or wt, after 2 weeks on 10 mg/l hygromycin.
DISCUSSION
We described here a set of mutations in Arabidopsis thaliana that interfere with epigenetic transgene silencing. The generation and identification of the mutants was based on a line containing linked multiple copies of a selectable resistance marker as a chromosomal insert. This initially expressed transgenic locus had undergone progressive epigenetic silencing in the course of seed amplification (13, 14). The failure to express hygromycin resistance in this line was not a result of a mutation in the hpt locus itself, because the locus could be reactivated at a low frequency under inductive conditions, such as a change of ploidy (14). The S5 generation was used for mutagenesis, and no spontaneous reactivation was observed among 74,000 M2 individuals examined. Several lines of evidence suggested that silencing occurred at the transcriptional level: silencing is meiotically stable (14) and associated with local hypermethylation and regional heterochromatization (O.M.S., unpublished data), analogous to transcriptional silencing of a different transgenic multicopy locus of Arabidopsis (22). These features made this Arabidopsis strain, containing the silent antibiotic resistance gene, an ideal material for a positive selection of mutations that release the silent state, resulting in antibiotic-resistant individuals. Such phenotypes are different from a class of previously described silencing mutants (12), which are more effective in transgene silencing than the wild type, mainly due to posttranscriptional inactivation.
Prior to the screen for mutants deficient in the maintenance of silencing, two potential problems were considered: (i) the majority of such mutations might be lethal and only recovered in the case of events with low penetrance, and (ii) some mutations might not cause immediate reactivation of the resistance marker in the selection-screened M2 generation, and under these conditions only null mutations would be recovered. In an attempt to mitigate these problems, we used two types of mutagenic treatment: incubation with EMS to induce mainly point mutations and irradiation with fast neutrons to provoke mainly large DNA rearrangements (23). The frequency of mutant recovery induced by EMS was 1 per 16,000 M2 individuals, similar to that described for the egs mutations (12). Unexpectedly, the FNR treatment yielded a higher rate of putative mutants (1 per 5,700), suggesting that rapid return to the active state was preferentially achieved by more severe alterations of trans-acting loci. More pronounced resistance and higher transcript levels obtained after FNR, rather than after EMS treatment, supported this hypothesis. Mutations identified by transgene reactivation had no consistent effects on the morphology and growth of the plants. This suggests that the mutations were not affecting essential genes.
The transmission of antibiotic resistance to selfed progeny of the seven self-fertile lines analyzed confirmed that the trait was heritable. However, backcrosses to the wild type resulted in F1 hybrids with phenotypes ranging from complete sensitivity to partial resistance (data not shown). Recessive mutations should not yield resistance in the F1, and dominant mutations should result in resistance of 100% or 50% of seedlings with som mutants being homozygous and hemizygous for the transgene, respectively. The variable ratios could be the result of dominant mutations with incomplete penetrance or of recessive mutations and delayed resilencing after the backcross. The second explanation is more convincing, because the progressive nature of silencing was well documented in another (24) and our system (14). Therefore, a reactivated, once-silent locus could undergo a gradual resilencing process. Such a partially active locus loses its value as an indicator of trans-acting silencing mutations and impedes further genetic analysis. This, together with the possibility of interaction between silent and active alleles, made it crucial to separate a putative mutation from the resistance locus once used and to reexamine the trans-activation ability of the mutations upon a clearly silent test locus. Trans-activation of such a test locus after a cross to a transgene-free mutant line also excluded that gene activity in the putative mutants resulted from an alteration within the transgene itself. Furthermore, it was a prerequisite for determining the genetic transmission of the mutation and the evaluation of its dominance. We performed such an assay for all mutant candidates after eliminating the residing resistance gene by segregation. The complete sensitivity of F1 hybrids between the transgene-free putative mutants and a parent with a silent test locus documented the recessive nature of the mutations. The mutant character and the independence from the transgene used for mutant selection was verified by F2 analysis, which revealed restored gene expression close to the value expected for the presence of trans-acting, single, and recessive mutations. This test cross was performed on the silent A locus. The transactivation ability of som mutants on other silent genes needs to be examined.
Several types of genes could have been encoded by SOM loci. Because transcriptional silencing is correlated with local changes in DNA methylation, it can be expected that some som mutations affect elements of this process. Arabidopsis strains (ddm) with a reduced level of methylation in repetitive DNA have been obtained in a mutant screen (20, 25) and by transgenic antisense suppression of a methyltransferase gene (METIas) (21, 26). We have observed that ddm1, but not METIas, showed a reactivating influence on the hpt locus, similar to som alleles. Methylation levels of the reactivated locus and of chromosomal repeats in som mutants were reduced to the same extend as in the genetic background of ddm1. The results of the complementation crosses suggested that three som mutations are allelic to ddm1. The failure of the METIas genotype to reverse activity of the silent hpt locus indicated that reduced activity of methyltransferase itself was probably not sufficient for the reactivation and unlikely to be the basis of a som mutation. Therefore, we expect that the som mutations not allelic to ddm1 or not yet assorted could affect other functions like chromatin components, chromatin modification, recognition of sequence duplication, or progression through the cell cycle.
The results of the allelism test with ddm1 and among the different soms are still ambiguous due to the unexpected parental interference with marker gene activity in F1. The reset of epigenetic states in both directions may require longer time and several generations. Such progressive resetting can cause complications and delay the definite genetic evidence; however, this is an intrinsic feature of epigenetics itself. Therefore, the genetic approach to transgene silencing may require a more sophisticated and laborious experimental setup, but in spite of these possible pitfalls, we still believe that a further characterization of mutants, like those presented here, should contribute substantially to our understanding of this phenomenon at the molecular level.
Acknowledgments
We thank Drs. Barbara Hohn, Thomas Hohn, Patrick King, and Frederick Meins for helpful comments on the manuscript and Paolo Amedeo and Michael Rothnie for their help in preparing the figures. We also thank Dr. H. Brunner from the IAEA laboratories in Seibersdorf, Austria, for the fast neutron irradiation, and Drs. Jean Finnegan and Eric Richards for kindly providing their plant material. We acknowledge financial support from the Swiss Federal Office for Education and Science (BBW Grant 93.0399–1/EU Grant CHRXCT940530 and BBW Grant 96.0250–1/EU Grant BIO4CT960253).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: hpt, hygromycin phosphotransferase; EMS, ethylmethane sulfonate; FNR, fast neutron irradiation; wt, wild type.
References
- 1.Meyer, P., ed. (1995) Curr. Top. Microbiol. Immunol. 197. [DOI] [PubMed]
- 2.Flavell R B. Proc Natl Acad Sci USA. 1994;91:3490–3496. doi: 10.1073/pnas.91.9.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzke M, Matzke A J M. Trends Genet. 1995;11:1–3. doi: 10.1016/s0168-9525(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 4.Matzke M, Primig M, Trnovsky J, Matzke A J M. EMBO J. 1989;8:643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer P, Saedler H. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:23–48. doi: 10.1146/annurev.arplant.47.1.23. [DOI] [PubMed] [Google Scholar]
- 6.Stam M, Mol J N M, Kooter J M. Ann Bot. 1997;79:3–12. [Google Scholar]
- 7.Singer M J, Selker E U. Curr Top Microbiol Immunol. 1995;197:165–177. doi: 10.1007/978-3-642-79145-1_11. [DOI] [PubMed] [Google Scholar]
- 8.Rossignol J-L, Faugeron G. Curr Top Microbiol Immunol. 1995;197:179–191. doi: 10.1007/978-3-642-79145-1_12. [DOI] [PubMed] [Google Scholar]
- 9.Henikoff S. Curr Top Microbiol Immunol. 1995;197:193–208. doi: 10.1007/978-3-642-79145-1_13. [DOI] [PubMed] [Google Scholar]
- 10.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:19–48. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 11.Reuter G, Spierer P. BioEssays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 12.Dehio C, Schell J. Proc Natl Acad Sci USA. 1994;91:5538–5542. doi: 10.1073/pnas.91.12.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mittelsten Scheid O, Paszkowski J, Potrykus I. Mol Gen Genet. 1991;228:104–112. doi: 10.1007/BF00282454. [DOI] [PubMed] [Google Scholar]
- 14.Mittelsten Scheid O, Jakovleva L, Afsar K, Maluszynska J, Paszkowski J. Proc Natl Acad Sci USA. 1996;93:7114–7119. doi: 10.1073/pnas.93.14.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masson J, Paszkowski J. Plant J. 1992;2:829–833. [Google Scholar]
- 16.Karesch H, Bilang R, Potrykus I. Plant Cell Rep. 1991;9:575–578. doi: 10.1007/BF00232335. [DOI] [PubMed] [Google Scholar]
- 17.Mittelsten Scheid O, Afsar K, Paszkowski J. Mol Gen Genet. 1994;244:325–330. doi: 10.1007/BF00285461. [DOI] [PubMed] [Google Scholar]
- 18.Klimyuk V, Carroll B J, Thomas C M, Jones J D G. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 19.Assaad F F, Tucker K L, Signer E R. Plant Mol Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- 20.Vongs A, Kakutani T, Martienssen R A, Richards E J. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 21.Finnegan E J, Peacock W J, Dennis E S. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye F, Signer E R. Proc Natl Acad Sci USA. 1996;93:10881–10886. doi: 10.1073/pnas.93.20.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldmann K A, Malmberg R L, Dean C. In: Arabidopsis. Meyerowitz E M, Somerville C R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 137–172. [Google Scholar]
- 24.Kilby N J, Leyser H M O, Furner I J. Plant Mol Biol. 1992;20:103–112. doi: 10.1007/BF00029153. [DOI] [PubMed] [Google Scholar]
- 25.Kakutani T, Jeddeloh J A, Flowers S K, Munakata K, Richards E J. Proc Natl Acad Sci USA. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronemus M J, Galbiati M, Ticknor C, Chen J, Dellaporta S L. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]